the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
The biogeography of relative abundance of soil fungi versus bacteria in surface topsoil
Kailiang Yu
Johan van den Hoogen
Zhiqiang Wang
Colin Averill
Devin Routh
Gabriel Reuben Smith
Rebecca E. Drenovsky
Kate M. Scow
Fei Mo
Mark P. Waldrop
Yuanhe Yang
Weize Tang
Franciska T. De Vries
Richard D. Bardgett
Peter Manning
Felipe Bastida
Sara G. Baer
Elizabeth M. Bach
Carlos García
Qingkui Wang
Linna Ma
Baodong Chen
Xianjing He
Sven Teurlincx
Amber Heijboer
James A. Bradley
Thomas W. Crowther
Fungi and bacteria are the two dominant groups of soil microbial communities worldwide. By controlling the turnover of soil organic matter, these organisms directly regulate the cycling of carbon between the soil and the atmosphere. Fundamental differences in the physiology and life history of bacteria and fungi suggest that variation in the biogeography of relative abundance of soil fungi versus bacteria could drive striking differences in carbon decomposition and soil organic matter formation between different biomes. However, a lack of global and predictive information on the distribution of these organisms in terrestrial ecosystems has prevented the inclusion of relative abundance of soil fungi versus bacteria and the associated processes in global biogeochemical models. Here, we used a global-scale dataset of >3000 distinct observations of abundance of soil fungi versus bacteria in the surface topsoil (up to 15 cm) to generate the first quantitative and high-spatial-resolution (1 km2) explicit map of soil fungal proportion, defined as fungi/fungi + bacteria, across terrestrial ecosystems. We reveal striking latitudinal trends where fungal dominance increases in cold and high-latitude environments with large soil carbon stocks. There was a strong nonlinear response of fungal dominance to the environmental gradient, i.e., mean annual temperature (MAT) and net primary productivity (NPP). Fungi dominated in regions with low MAT and NPP and bacteria dominated in regions with high MAT and NPP, thus representing slow vs. fast soil energy channels, respectively, a concept with a long history in soil ecology. These high-resolution models provide the first steps towards representing the major soil microbial groups and their functional differences in global biogeochemical models to improve predictions of soil organic matter turnover under current and future climate scenarios. Raw datasets and global maps generated in this study are available at https://doi.org/10.6084/m9.figshare.19556419 (Yu, 2022).
- Article
(3734 KB) - Full-text XML
-
Supplement
(11481 KB) - BibTeX
- EndNote
Fungi and bacteria are the dominant members of soil microbial communities worldwide in terms of diversity, abundance and biomass (Bahram et al., 2018). Representing distinct kingdoms of life, bacteria and fungi systematically differ in a multitude of physiological and life-history traits with direct implications for global soil biogeochemical cycles (Waring et al., 2013; Wieder et al., 2015), including the decomposition of organic matter, which contributes to one of the largest fluxes of CO2 on earth (Glassman et al., 2018). Compared to bacteria, fungi generally have slower growth and turnover rates (Rousk and Bååth, 2007), a greater carbon (C) to nutrient stoichiometry (Waring et al., 2013), a greater capacity to degrade a wider and more recalcitrant range of substrates (Strickland and Rousk, 2010) and, potentially, a higher C use efficiency (Soares and Rousk, 2019). For these reasons, a new generation of soil and ecosystem models have begun to explicitly represent these fundamentally distinct fast and slow cycling microbial groups, suggesting that spatially explicit information about the relative abundance of fungi versus bacteria in a region can dramatically improve the accuracy of global carbon cycling model predictions (Shi et al., 2018; Sulman et al., 2014; Wieder et al., 2013, 2015). Generating an understanding of the factors affecting the biogeography of the relative abundance of fungi versus bacteria in soil, and its connection to the global carbon cycle, would represent a breakthrough step forward in our general understanding of the natural history of soil microbial life.
Temperature, precipitation, soil pH and soil C:N have all been linked to the balance of fungi vs. bacteria within soil communities across different spatial scales (Bahram et al., 2018; Strickland and Rousk, 2010; Tedersoo et al., 2014). Relative to fungi, bacteria tend to dominate in locations with high soil nutrient contents or in frequently disturbed soils that limit the growth of fungal hyphae or make N more available (Fierer et al., 2009; Van Der Heijden et al., 2008; Strickland and Rousk, 2010). However, until now, the relative importance of these different environmental drivers at the global scale remains relatively unclear, and the biogeography of these major functional groups (fungi vs. bacteria) has only been demonstrated at local and regional scales. A recent analysis suggested that the relative soil bacterial abundance is high in tropical latitudes and decreases in abundance towards the high-latitude boreal regions, where fungi tend to dominate (Bahram et al., 2018). Translating these broad-scale trends into quantitative, spatially explicit information will be necessary if we intend to represent regional variations in soil community functioning (Wieder et al., 2013, 2015) or predict future changes in terrestrial carbon and nutrient cycling.
Some progress was made in the quantitative and spatially explicit understanding of global biogeographic patterns of fungal and bacterial biomass and their biomass ratio. By synthesizing phospholipid-derived fatty acid data from 1323 locations across the globe and extrapolating linear relationships with environmental factors, a recent study generated global maps of fungal and bacterial biomass and their biomass ratio at a resolution of 0.5∘ for topsoil (0–30 cm) (He et al., 2020). This approach provided support for the broad-scale latitudinal trends, with a high fungal dominance in high-latitude regions. Yet, there are still three crucial knowledge gaps to address. First, we still lack a high-resolution evaluation of the spatial patterns and regional contingencies in fungal : bacterial ratios, which would allow the representation of microbe-mediated mechanisms that operate within and/or across ecosystems at fine scales (Frindte et al., 2019; Zhu et al., 2017). Second, the response of soil microbial community composition across environmental gradients is expected to be nonlinear, with strong interactive effects of different environmental characteristics that give rise to thresholds that diverge from the global latitudinal trends (Sengupta et al., 2021; Wang et al., 2018; Waring et al., 2013). This nonlinear linkage of soil microbial communities with the environmental resource gradient has not been assessed, even though it has fundamental implications for ecosystem functions and management solutions (Sengupta et al., 2021; Wang et al., 2018). Third, there are distinct differences in soil nutrients, the soil microbial community and the associated biogeochemical processes across soil depths, i.e., from surface topsoil (i.e., 0–10 cm) to subsurface topsoil (i.e., 0–30 cm) (Lavahun et al., 1996; Yue et al., 2015). A continental-scale empirical study further showed that strong positive associations among the soil microbial community, fertility and plant productivity are limited to the surface topsoil (Delgado-Baquerizo et al., 2017), thus highlighting the potentially dominant role of surface topsoil microbial communities in regulating ecosystem functions and the need for research that can provide a global spatially explicit understanding of soil fungi versus bacteria in surface topsoil.
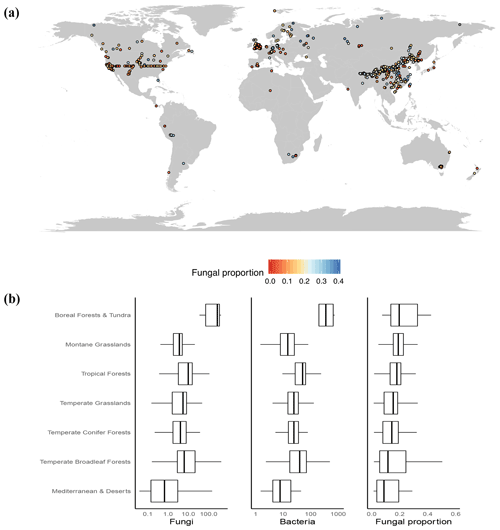
Figure 1(a) Map of sample locations and fungal proportion data. A total of 3224 samples were collected and aggregated into 943 1 km2 pixels that were used for geospatial modeling. (b) The medians and interquartile ranges of fungal abundance, bacterial abundance and fungal proportion in various vegetation biomes. Since they had low sample sizes (<25), tundra and boreal forest data were combined, as were Mediterranean and desert data.
Here, we present a global analysis of the total and relative abundances of soil fungi versus bacteria in soil surfaces (defined as the top 10–15 cm) informed by over 3000 spatially distinct surface soil observations of phospholipid-derived fatty acids (PLFAs) (Fig. 1a). The use of PLFA data provides an opportunity to provide quantitative insights into the abundances of the major functional groups. We conducted the analysis of the abundances in view of the uncertainty in conversion factors used to convert the abundance derived from PLFA to biomass (Frostegård et al., 2011; Klamer and Bååth, 2004). We used machine learning to link the variation in soil fungi versus bacteria abundances to the global variation in 95 climate, vegetation and soil variables. This allowed us to (1) explore the environmental drivers of fungal and bacterial dominance, defined as fungal proportion, i.e., fungi (fungi + bacteria), where values closer to 1 indicate a higher fungal dominance and values closer to zero indicate a greater bacterial dominance (see the “Methods” section), and (2) examine the nonlinear response or pattern of fungal proportion across environmental gradients, i.e., mean annual temperature (MAT) and net primary productivity (NPP). Based on the observed relationships (accounting for the nonlinearity), we generated a quantitative spatially explicit global map (1 km2) of fungal proportion and assessed how soil fungal and bacterial dominance varies with key climate, soil, vegetation and geographic drivers.
2.1 Data acquisition of soil microbe composition
We compiled data on the abundance of soil fungi versus bacteria and the fungal proportion, defined as fungi (fungi + bacteria). We focused on phospholipid-derived fatty acids (PLFAs), and the data derived from PLFAs regarding the balance between fungal and bacterial PLFAs (Frostegård et al., 2011) can provide a valuable estimation of the comparative dominance of both functional groups. Data based on qPCR were not included because of the difference in units with PLFAs. With nonsignificant difference of the general pattern and conclusion using data on fungal proportion and the fungi : bacteria ratio, we focused on and reported the results on the fungal proportion rather than the fungi : bacteria ratio because (1) the fungal proportion is insensitive to whether fungi or bacteria are the numerator (i.e., bacterial proportion fungal proportion) and (2) the spread of the frequency distribution of the fungal proportion was greater and thus led to better machine learning predictions (Fig. S1 in the Supplement). The data were compiled in a primary literature review performed through Google Scholar, Web of Science (http://apps.webofknowledge.com, last access: 30 June 2020) and the China National Knowledge Infrastructure Database (http://cnki.net, last access: 30 June 2020) up to 30 June 2020 using the keywords “fungi”, “bacteria”, “abundance” and “PLFA”. To be included in our data analysis, the study had to at least have the following metadata: longitude and latitude, sampling date, sampling depth, information on land use (agriculture, tree plantations, or natural sites), units and the methods used. In total, this led to 319 references. We further used the following criteria to select eligible references and datasets. (1) When the studies were manipulative experiments, we only included the data from “control” plots (Chen et al., 2016). (2) We standardized our efforts by focusing on all samples that were collected from surface topsoils (≈0–10/15 cm) because this layer contains the greatest biomass and the majority of samples were taken from it. (3) We used the datasets that reported abundance in units of nmol, µmol, or mol %, since the majority (>90 %) of the datasets reported abundance. Thus, we excluded all datasets that reported biomass instead of abundance. (4) We excluded observations located in the sea, since our study focuses on terrestrial ecosystems. (5) We only included the datasets on soil samples derived from field experiments and thus excluded the datasets from incubation experiments. (6) Some datasets reported in the original references as averages across sampling sites or sampling dates were included.
The criteria were carefully scrutinized by three independent researchers, and this ultimately led to the use of 179 eligible references (see the references for PLFAs in the Supplement) in this study. In total, we compiled a dataset of fungal proportion (n=3224) at a global scale. The subset of data (n=1795) on only natural ecosystems (Fig. S2a in the Supplement) were used to examine the potential role of land-use change (see the “Methods” section in the Supplement). The results showed a minimal difference between the two scenarios of including all data and including only data on natural ecosystems. All data points falling within the same 30 arcsec (∼1 km2) pixel were aggregated via an average. The aggregated data on fungal proportion (n=946 for all data; n=716 for natural ecosystems) were used to examine its environmental controls and in geospatial modeling to create the global map (Figs. 1a and S2a).
The spatial variations of the fungi to bacteria ratio or fungal proportion across latitude could be influenced by changes (increases or decreases) in either the abundance of fungi, the abundance of bacteria, or both. Thus, to better understand the biogeographic pattern of fungal and bacterial composition, we also analyzed the spatial patterns of abundance of fungi and bacteria by using abundance data with the same unit (nmol g−1 PLFA). In total, compiling our data led to final subsets of 2753 and 2759 samples which were used for further analyses of the abundances of fungi and bacteria, respectively (Fig. S3). As compared to the larger sample size of fungal proportion (n=946 for all data), data on the abundances of fungi (n=646 for all data) and bacteria (n=647 for all data) aggregated within a 30 arcsec (∼1 km2) pixel via an average were used for the analysis of their spatial trends across vegetation biomes, vegetation types and latitudes (see the “Methods” section in the Supplement).
2.2 Geospatial modeling
A stack (n=95) of ecologically relevant global map layers, including soil physical, chemical and nutrient properties, climate conditions, vegetative indices, radiation and topographic variables, and anthropogenic covariates (Table S1 in the Supplement), were used to derive the environmental factors which could affect fungal proportion. All of these covariate map layers were standardized at 30 arcsec resolution (≈1 km2 at the Equator) (van den Hoogen et al., 2019). These covariates were then derived based on the georeferenced coordinates of the soil samples aggregated at 30 arcsec resolution.
We used a random forest machine learning algorithm (see the “Methods” section in the Supplement) with the derived 95 covariates to extrapolate these relationships between fungal proportion and environmental conditions across the globe and generated the first spatially explicit quantitative map of fungal proportion at a global scale. The strength of prediction was evaluated using k-fold cross-validation (with k=10) and the best models with a high coefficient of determination and a low standard deviation in the mean cross-validation were used to generate the global map of fungal proportion. The standard error sharply decreased with increasing sample size across all vegetation biomes, and the analysis showed that an efficient prediction required a large sample size (n>500) (Fig. S4). To evaluate the sensitivity, we also generated a map of uncertainty (standard deviation as a fraction of the mean predicted value) of fungal proportion by using a stratified bootstrapping procedure (van den Hoogen et al., 2019). The fungal proportion data were stratified by biome to avoid biases. In total, 100 bootstrap iterations were used, thus generating 100 global maps of fungal proportion that were used to quantify statistically robust 95 % confidence intervals per pixel.
2.3 Environmental drivers and statistic analysis
To examine the environmental controls of soil microbial composition at a global scale, we chose the top drivers (Chen et al., 2016; Drenovsky et al., 2010a; de Vries et al., 2012), which include soil properties, climate conditions, vegetation index and human activities (see the “Methods” section in the Supplement). These variables were examined to avoid multicollinearity: a matrix of pairwise correlations was used to remove any variable with high correlations (R>0.7) with other predictor variables (Anderegg et al., 2013). A random forest machine learning algorithm was then used to determine the importance of each variable (Breiman, 2001). The mean decrease in accuracy (% IncMSE) and the mean decrease in the Gini coefficient (IncNodePurity) were reported, and the variables with greater values of % IncMSE and IncNodePurity are more important in influencing fungal proportion. Partial functions of the most important variables (MAT and NPP) were plotted using the forestFloor package to examine their influences on fungal proportion.
3.1 Raw data patterns of fungal proportion
Globally, we observed a greater than 10-fold variation in soil fungal proportion across all sites, ranging from 0.01 to 0.6 (Fig. 1b). At a global scale, we found clear latitudinal trends, with both the abundance of fungi and the abundance of bacteria increasing in high-latitude regions. Yet, the abundance of fungi increased with latitude at a greater rate than the abundance of bacteria (Fig. S5), resulting in a higher proportion of fungi in the cold, high-latitude regions. These latitudinal trends lend support to the general global patterns detected in a previous broad-scale analysis (Bahram et al., 2018) and in a recent meta-data analysis (He et al., 2020). As such, the highest fungal dominance was observed in tundra and boreal forest ecosystems (mean ±1 SE: 0.23±0.02; Fig. 1b). In addition, highly elevated and cold grasslands (i.e., montane grasslands) with a large soil organic C (SOC) content generally harbor a higher proportion of fungi relative to bacteria (Fig. 1b).
In similar climates, the abundances of soil fungi versus bacteria as well as the fungal proportion were greatest in ecosystems harboring woody vegetation compared to grasslands and managed (agricultural) ecosystems (Fig. S6). This finding is consistent with the idea that ecosystems dominated by woody plants generate lignified, more recalcitrant and nutrient-poor soil C inputs that characteristically favor fungal dominance (Fierer et al., 2009; Strickland and Rousk, 2010) and have a biomass stoichiometry that is better suited to low-nutrient environments (Waring et al., 2013). But we stress that this link of belowground soil microbial composition (fungi vs. bacteria) with aboveground plant community composition (woody plants vs. grasses) can be complex, nonlinear and even divergent, as demonstrated by the well-mixed fungi vs. bacteria abundances in both grasslands and forests but the nonexistence of woody plants in grasslands and scarcity of grasses in forests. This raises our curiosity as to whether the interactions, associations, or couplings of belowground soil microbial composition with aboveground plant community composition are stronger in ecosystems where woody plants and grasses interact or coexist (i.e., savannas) (Yu and D'Odorico, 2015). It also remains unclear how this coupling could improve our understanding of ecosystem carbon cycling and other services.
The management of agricultural ecosystems often disrupts soil fungal networks (i.e., tillage, frequent dry/wet cycles due to irrigation, machine operations, etc.), which decreases the abundance of fungi relative to bacteria in agricultural soils (Fig. S6) (Drenovsky et al., 2010b; Jangid et al., 2011; Waldrop et al., 2017). A central concern in agricultural ecosystems is the tradeoff of increased food production to feed the increasing population vs. the decreased soil carbon storage, which accelerates global climate change (Sanderman et al., 2017). This study shows a higher bacterial abundance relative to fungal abundance in soils of agricultural lands where soil carbon storage is low; this corresponds with the global trend for bacterial dominance at low latitudes where soil carbon storage is low. These results suggest potential strong but complex interactions and feedbacks between soil microbial composition and soil functions (i.e., soil carbon storage) (Bardgett et al., 2008), while the mechanistic links need further studies.
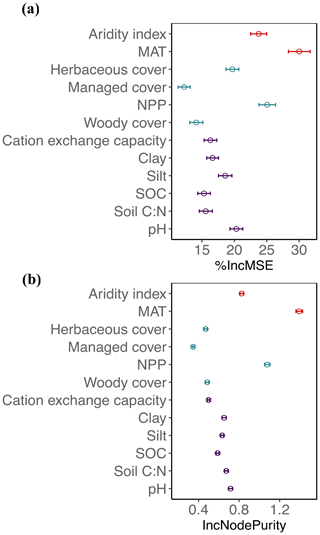
Figure 2Mean decrease in accuracy (% IncMSE, mean and SD; a) and mean decrease in the Gini coefficient (IncNodePurity, mean and SD; b) estimated from 1000 simulations of random forests. This was used to evaluate the importance of top environmental drivers in the proportion of fungi derived from the “all” dataset.
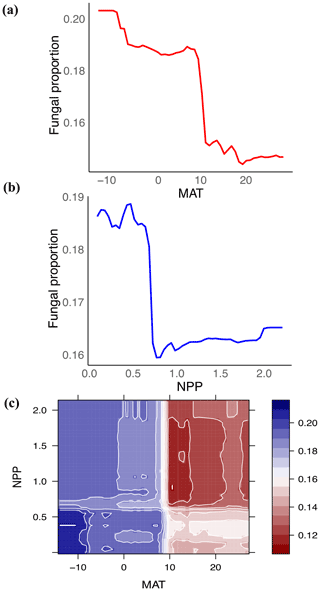
Figure 3Fungal proportion is primarily associated with climate: mean annual temperature (MAT) and net primary productivity (NPP). (a, b) Partial feature contributions of the primary environmental variables (a) MAT and (b) NPP to fungal proportion. (c) Partial feature contributions of primary environmental variable interactions (MAT vs. NPP) to fungal proportion.
3.2 Drivers of fungal proportion
Globally, the fungal proportion in soil can be predicted by a few primary environmental drivers (Figs. 2 and S7). Specifically, mean annual temperature (MAT) and primary productivity (NPP) were found to be strong determinants of fungal dominance. The responses of fungal proportion to both MAT and NPP were strongly nonlinear, with warmer, more productive regions of the world (i.e., tropical forest biomes) showing a lower dominance of fungi as compared to colder, less productive ecosystems (i.e., boreal forest and tundra biomes, Figs. 3 and S8). This pattern is consistent with the idea that fungi and bacteria represent slow vs. fast soil energy channels, respectively (Crowther et al., 2019; Malik et al., 2016), a concept with a long history in soil ecology (Moore et al., 2003; Moore and William Hunt, 1988). This finding is important because it could potentially link the belowground slow (fungi) vs. fast (bacteria) energy channels with aboveground slow plant growth rates (woody plants) vs. fast growth rates (grasses), while the linkage could be complex, nonlinear or even divergent. The fast vs. slow concept or spectrum has fundamentally improved the understanding and prediction of land carbon storage across resource gradients or under global change. There could typically be a tradeoff between faster growth and higher mortality or heterotrophic respiration with resource (i.e., CO2)-enriched conditions (Jiang et al., 2020; Terrer et al., 2021; Yu et al., 2019), thus constraining land carbon storage. This raises the question of how the belowground fast vs. slow energy channels and the aboveground fast vs. slow growth spectrum could potentially be linked or integrated to assess land carbon storage.
Temperature can affect soil microbial composition in complex ways: directly via physiology or indirectly via the soil substrate (Romero-Olivares et al., 2017). Previous studies have shown a nonlinear response of the soil fungal and bacterial ratio to soil substrate nutrient availability (Waring et al., 2013). A nonlinear trend in the temperature sensitivity (Q10) of soil organic C decomposition, as regulated by the soil fungal and bacterial ratio, was also found with latitude (Wang et al., 2018). Other environmental variables such as the soil C to nitrogen ratio (C:N) have previously been found to be important drivers of fungal proportion within local- and regional-scale analyses (Fierer et al., 2009; Waring et al., 2013). Our results suggest a more complicated relationship between fungal proportion and the soil C:N. In the low range of soil C:N values, fungal proportion decreased with soil C:N (Fig. S9a), suggesting that the role of site-specific differences (i.e., climate or plant community) likely outweighs the influence of N availability (Soares and Rousk, 2019). Aside from these ecosystems, we observed a positive relationship between fungal proportion and soil C:N at a global scale, consistent with previous work at local and regional scales (Strickland and Rousk, 2010; Waring et al., 2013). Additionally, pH has been thought of as a critical driver of microbial diversity and biomass in soils. At local scales, previous studies have reported either no relationship, a negative correlation or a convex curve between the fungal and bacterial ratio and soil pH (Rousk et al., 2009, 2010; de Vries et al., 2012). Our global-scale analysis suggests a convex relationship between fungal proportion and soil pH, with fungi dominating only within a narrow pH range (<5–6) (Fig. S9b).
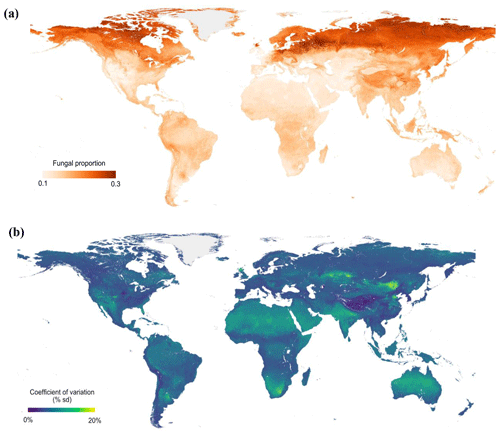
Figure 4Global maps of fungal proportion (a) and bootstrapped (100 iterations) coefficient of variation (b) at the 30 arcsec (approximately 1 km2) pixel scale. The bootstrapped coefficient of variation is the standard deviation divided by the mean predicted value and is a measure of prediction accuracy. Sampling was stratified by biome.
3.3 Biogeographic pattern from the machine learning model
Across all samples, the machine learning model was able to predict the variation in fungal and bacterial dominance with high predictive accuracy (R2=0.43/0.35 in 10-fold cross-validation, R2=0.92/0.91 in the final model; Fig. S10a and b). By extrapolating these relationships across terrestrial ecosystems, we could identify clear global trends in fungal dominance. Despite these general global-scale patterns of an increase in fungi dominance with latitude, our models also revealed regions that diverge from the global trends (Figs. 4a and S11a). For instance, northeastern Europe is dominated by woody vegetation and exhibits a high fungal proportion, while the United Kingdom and northern Kazakhstan have much lower fungal proportions despite being at comparable latitudes, likely because these areas are dominated by herbaceous vegetation with lower lignin contents than in woody tissues. Tibetan alpine grasslands are at much lower latitudes but have high values of fungal proportion, in part due to very high SOC stocks and cold temperatures. Model predictions of fungal proportion had high uncertainty in dry regions (i.e., northern and southern Africa, Australia, western USA and eastern Mongolia) (Figs. 4b and S11b), presumably because of the low sample size from drylands and/or the complex responses of fungi and bacteria to water availability (Fierer et al., 2009; Strickland and Rousk, 2010). Indeed, our datasets are mostly concentrated in the US, Europe and East Asia, thus highlighting the data gaps for tropical and boreal biomes. Even for the temperate biome, there were data gaps in West Australia and central Asia. Because of the unbalanced sample distribution, we also used a bootstrapping strategy (100 iterations) by randomly sampling 90 % of the data with replacement. The results showed similar spatial patterns of fungal proportion (Fig. S12a) and uncertainty (Fig. S12b) to those obtained using the full dataset without bootstrapping.
Our study differs from a previous study (He et al., 2020) in several aspects, including sample size (n>3000), spatial resolution (1 km2), consideration of nonlinearity (through random forest analysis) and soil depth (soil surface 0–10/15 cm). We also note that our analysis sticks to the original data on abundance derived from PLFA instead of converting abundance to biomass. The conversion of abundance to biomass needs the conversion factor, which has large uncertainty (Frostegård et al., 2011; Klamer and Bååth, 2004). Our high-resolution map allows the representation of microbe-mediated mechanisms at fine scales, and therefore allows us to link these mechanisms with ecosystem functions. For instance, the significant functional differences between fungi and bacteria mean that the relative dominance of fungi vs. bacteria is likely to influence a wide range of ecosystem functions such as C use efficiency (CUE) of the decomposer community (Six et al., 2006; Soares and Rousk, 2019) and enzymatic activity in soil N vs. P acquisition (Caldwell, 2005; Crowther et al., 2019). At fine, local or even regional scales, these relationships between soil microbial composition and ecosystem functions could only be identified well using fine-scale maps of soil microbial composition.
3.4 Implications and limitations of this study
It is generally accepted that the soil microbiome exerts major control over soil processes and in turn ecosystem functioning, and by extension the global biogeochemical cycles (Bahram et al., 2018; Crowther et al., 2019; Van Der Heijden et al., 2008; Jenny, 1941). Fungi and bacteria represent most of the diversity of life on earth (Bardgett and van der Putten, 2014; Locey and Lennon, 2016). Yet, the inclusion of fungal and bacterial abundances in quantitative ecosystem and earth system models has been hindered by the paucity of information about organisms at appropriate spatial scales. Here, we impose a global top-down constraint on the broad composition of soil microbial life. By doing so, we hope to empower microbial, ecosystem and earth-system scientists to consider how this broad constraint on the soil biodiversity may inform and transform our understanding of terrestrial ecosystem functioning. As we develop a spatially explicit understanding of the global soil community, we will be able to better parameterize and benchmark our predictions about the rate and efficiency of carbon turnover in soil and the feedbacks to ongoing climate change.
Despite the progress made in this study, we must clarify two limitations of this study. First, our study highlights the data gaps in fungal proportion prediction at low latitudes (tropical biome) and high latitudes (boreal biome, i.e., boreal forests and tundra). Tropical and boreal biomes are hotspots or debated regions in terms of their relative capacity and capability to sequestrate atmospheric CO2 and mitigate climate change in an increasingly changing climate (Schimel et al., 2015; Tagesson et al., 2020). They are also regions with striking differences in soil microbial composition (fungal proportion), plant communities and soil carbon storage, thus suggesting that there are potentially strong interactions and feedbacks between these factors in these regions (Bardgett et al., 2008). The boreal biome contains a large amount of soil organic carbon, which could be sensitive to global change (i.e., warming), wherein the soil microbial community (i.e., the total biomass or the relative abundance of soil fungi versus bacteria) could play an essential role. Second, microbial biomass (C) is more relevant to the soil carbon cycling and carbon stock due to its contribution to living carbon pools and the impacts of its microbial necromass (Liang et al., 2019), while the conversion factor for the conversion of abundance into biomass is currently not available. To mechanistically and explicitly incorporate soil microbial composition into biogeochemical models, the biogeographic pattern of abundance or biomass of each major group (fungi vs. bacteria) and the relative ratios of categories of fungi (i.e., saprotrophic fungi, arbuscular mycorrhizal fungi vs. ectomycorrhizal fungi) and/or bacteria (i.e., gram-positive bacteria vs. gram-negative bacteria) would also be critical, in view of their striking functional difference (Averill et al., 2014; Crowther et al., 2019). These research gaps highlight the urgent need for new research that utilizes the increasing availability of datasets.
The map and data for this manuscript are available at https://doi.org/10.6084/m9.figshare.19556419 (Yu, 2022).
This study used a global-scale dataset of >3000 distinct observations of abundance of soil fungi versus bacteria in the surface topsoil (up to 15 cm) to generate the first quantitative and high-spatial-resolution (1 km2) explicit maps of relative abundance of soil fungi versus bacteria across global terrestrial ecosystems. Our machine learning approach (random forest) enabled us to link the variation in fungal proportion to global variations in climate, soil, vegetation and other environmental drivers whilst accounting for the interactions and nonlinearities among them. We found striking latitudinal trends where fungal dominance increases in cold and high-latitude environments with large soil carbon stocks. The fungal proportion in soil can be predicted by a few primary environmental drivers: temperature and NPP, which show strong nonlinear effects. We demonstrated that fungi and bacteria represent slow vs. fast energy channels, whereby they dominate in regions of low MAT and NPP vs. high MAT and NPP, respectively. Overall, our spatially explicit model would enable us to explicitly represent the different contributions of fast (bacterial) vs. slow (fungal) energy channels in spatially explicit biogeochemical models, with the potential to enhance the accuracy of soil carbon turnover and carbon storage predictions. We further highlight the data gaps in tropical and boreal regions and the need for future research endeavors to generate high-resolution biogeographic patterns of the biomass of each major microbial group and the relative biomass ratios between and within major microbial groups.
The supplement related to this article is available online at: https://doi.org/10.5194/essd-14-4339-2022-supplement.
KY and TWC designed the project. KY built the PLFA datasets with help from JvdH and ZW. KY performed the analysis with inputs from DR and CA. KY, CA and TWC wrote the paper, with revisions from all other coauthors. GRS, RED, KMS, FM, MPW, YY, FTDV, RDB, PM, FB, SGB, EMB, CG, QW, LM, BC, XH, WT, ST, AH and JAB contributed to the PLFA datasets.
The contact author has declared that none of the authors have any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
We are grateful for the data contributors and the scientific community which made the data accessible and useful for our study. Zhiqiang Wang would like to acknowledge the funding support by International Science and Technology Cooperation Project of Qinghai province of China (2022-HZ-817).
This research has been supported by T.W.C. from DOB ecology.
This paper was edited by David Carlson and reviewed by two anonymous referees.
Anderegg, L. D. L., Anderegg, W. R. L., Abatzoglou, J., Hausladen, A. M., and Berry, J. A.: Drought characteristics' role in widespread aspen forest mortality across Colorado, USA, Glob. Chang. Biol., 19, 1526–1537, https://doi.org/10.1111/gcb.12146, 2013.
Averill, C., Turner, B. L., and Finzi, A. C.: Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage, Nature, 505, 543–545, https://doi.org/10.1038/nature12901, 2014.
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., Bengtsson-Palme, J., Anslan, S., Coelho, L. P., Harend, H., Huerta-Cepas, J., Medema, M. H., Maltz, M. R., Mundra, S., Olsson, P. A., Pent, M., Põlme, S., Sunagawa, S., Ryberg, M., Tedersoo, L., and Bork, P.: Structure and function of the global topsoil microbiome, Nature, 560, 233–237, https://doi.org/10.1038/s41586-018-0386-6, 2018.
Bardgett, R. D. and van der Putten, W. H.: Belowground biodiversity and ecosystem functioning., Nature, 515, 505–511, https://doi.org/10.1038/nature13855, 2014.
Bardgett, R. D., Freeman, C., and Ostle, N. J.: Microbial contributions to climate change through carbon cycle feedbacks, ISME J., 2, 805–814, https://doi.org/10.1038/ismej.2008.58, 2008.
Breiman, L.: Random forests, Mach. Learn., 45, 5–32, https://doi.org/10.1023/A:1010933404324, 2001.
Caldwell, B. A.: Enzyme activities as a component of soil biodiversity: A review, Pedobiologia, 49, 637–644, https://doi.org/10.1016/j.pedobi.2005.06.003, 2005.
Chen, Y. L., Ding, J. Z., Peng, Y. F., Li, F., Yang, G. B., Liu, L., Qin, S. Q., Fang, K., and Yang, Y. H.: Patterns and drivers of soil microbial communities in Tibetan alpine and global terrestrial ecosystems, J. Biogeogr., 43, 2027–2039, https://doi.org/10.1111/jbi.12806, 2016.
Crowther, T. W., van den Hoogen, J., Wan, J., Mayes, M. A., Keiser, A. D., Mo, L., Averill, C., and Maynard, D. S.: The global soil community and its influence on biogeochemistry, Science, 365, eaav0550, https://doi.org/10.1126/science.aav0550, 2019.
Delgado-Baquerizo, M., Powell, J. R., Hamonts, K., Reith, F., Mele, P., Brown, M. V., Dennis, P. G., Ferrari, B. C., Fitzgerald, A., Young, A., Singh, B. K., and Bissett, A.: Circular linkages between soil biodiversity, fertility and plant productivity are limited to topsoil at the continental scale, New Phytol., 215, 1186–1196, https://doi.org/10.1111/nph.14634, 2017.
de Vries, F. T., Manning, P., Tallowin, J. R. B., Mortimer, S. R., Pilgrim, E. S., Harrison, K. A., Hobbs, P. J., Quirk, H., Shipley, B., Cornelissen, J. H. C., Kattge, J., and Bardgett, R. D.: Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities, Ecol. Lett., 15, 1230–1239, https://doi.org/10.1111/j.1461-0248.2012.01844.x, 2012.
Drenovsky, R. E., Steenwerth, K. L., Jackson, L. E., and Scow, K. M.: Land use and climatic factors structure regional patterns in soil microbial communities, Glob. Ecol. Biogeogr., 19, 27–39, https://doi.org/10.1111/j.1466-8238.2009.00486.x, 2010a.
Drenovsky, R. E., Steenwerth, K. L., Jackson, L. E., and Scow, K. M.: Land use and climatic factors structure regional patterns in soil microbial communities, Glob. Ecol. Biogeogr., 19, 27–39, https://doi.org/10.1111/j.1466-8238.2009.00486.x, 2010b.
Fierer, N., Strickland, M. S., Liptzin, D., Bradford, M. A., and Cleveland, C. C.: Global patterns in belowground communities, Ecol. Lett., 12, 1238–1249, https://doi.org/10.1111/j.1461-0248.2009.01360.x, 2009.
Frindte, K., Pape, R., Werner, K., Löffler, J., and Knief, C.: Temperature and soil moisture control microbial community composition in an arctic–alpine ecosystem along elevational and micro-topographic gradients, ISME J., 13, 2031–2043, https://doi.org/10.1038/s41396-019-0409-9, 2019.
Frostegård, Å., Tunlid, A., and Bååth, E.: Use and misuse of PLFA measurements in soils, Soil Biol. Biochem., 43, 1621–1625, https://doi.org/10.1016/j.soilbio.2010.11.021, 2011.
Glassman, S. I., Weihe, C., Li, J., Albright, M. B. N., Looby, C. I., Martiny, A. C., Treseder, K. K., Allison, S. D., and Martiny, J. B. H.: Decomposition responses to climate depend on microbial community composition, P. Natl. Acad. Sci. USA, 115, 11994–11999, https://doi.org/10.1073/pnas.1811269115, 2018.
He, L., Mazza Rodrigues, J. L., Soudzilovskaia, N. A., Barceló, M., Olsson, P. A., Song, C., Tedersoo, L., Yuan, F., Yuan, F., Lipson, D. A., and Xu, X.: Global biogeography of fungal and bacterial biomass carbon in topsoil, Soil Biol. Biochem., 151, 108024, https://doi.org/10.1016/j.soilbio.2020.108024, 2020.
Jangid, K., Williams, M. A., Franzluebbers, A. J., Schmidt, T. M., Coleman, D. C., and Whitman, W. B.: Land-use history has a stronger impact on soil microbial community composition than aboveground vegetation and soil properties, Soil Biol. Biochem., 43, 2184–2193, https://doi.org/10.1016/j.soilbio.2011.06.022, 2011.
Jenny, H.: Factors of Soil Formation, Soil Sci., 52, 415, https://doi.org/10.1097/00010694-194111000-00009, 1941.
Jiang, M., Medlyn, B. E., Drake, J. E., Duursma, R. A., Anderson, I. C., Barton, C. V. M., Boer, M. M., Carrillo, Y., Castañeda-Gómez, L., Collins, L., Crous, K. Y., De Kauwe, M. G., dos Santos, B. M., Emmerson, K. M., Facey, S. L., Gherlenda, A. N., Gimeno, T. E., Hasegawa, S., Johnson, S. N., Kännaste, A., Macdonald, C. A., Mahmud, K., Moore, B. D., Nazaries, L., Neilson, E. H. J., Nielsen, U. N., Niinemets, Ü., Noh, N. J., Ochoa-Hueso, R., Pathare, V. S., Pendall, E., Pihlblad, J., Piñeiro, J., Powell, J. R., Power, S. A., Reich, P. B., Renchon, A. A., Riegler, M., Rinnan, R., Rymer, P. D., Salomón, R. L., Singh, B. K., Smith, B., Tjoelker, M. G., Walker, J. K. M., Wujeska-Klause, A., Yang, J., Zaehle, S., and Ellsworth, D. S.: The fate of carbon in a mature forest under carbon dioxide enrichment, Nature, 580, 227–231, https://doi.org/10.1038/s41586-020-2128-9, 2020.
Klamer, M. and Bååth, E.: Estimation of conversion factors for fungal biomass determination in compost using ergosterol and PLFA 18:2ω6,9, Soil Biol. Biochem., 36, 57–65, https://doi.org/10.1016/j.soilbio.2003.08.019, 2004.
Lavahun, M. F. E., Joergensen, R. G., and Meyer, B.: Activity and biomass of soil microorganisms at different depths, Biol. Fertil. Soils, 23, 38–42, https://doi.org/10.1007/BF00335816, 1996.
Liang, C., Amelung, W., Lehmann, J., and Kästner, M.: Quantitative assessment of microbial necromass contribution to soil organic matter, Glob. Chang. Biol., 25, 3578–3590, https://doi.org/10.1111/gcb.14781, 2019.
Locey, K. J. and Lennon, J. T.: Scaling laws predict global microbial diversity, P. Natl. Acad. Sci. USA, 113, 5970–5975, https://doi.org/10.1073/pnas.1521291113, 2016.
Malik, A. A., Chowdhury, S., Schlager, V., Oliver, A., Puissant, J., Vazquez, P. G. M., Jehmlich, N., von Bergen, M., Griffiths, R. I., and Gleixner, G.: Soil fungal: Bacterial ratios are linked to altered carbon cycling, Front. Microbiol., 7, 1247, https://doi.org/10.3389/fmicb.2016.01247, 2016.
Moore, J. C. and William Hunt, H.: Resource compartmentation and the stability of real ecosystems, Nature, 333, 261–263, https://doi.org/10.1038/333261a0, 1988.
Moore, J. C., McCann, K., Setälä, H., and De Ruiter, P. C.: Top-down is bottom-up: Does predation in the rhizosphere regulate aboveground dynamics?, Ecology, 84, 846–857, https://doi.org/10.1890/0012-9658(2003)084[0846:TIBDPI]2.0.CO;2, 2003.
Romero-Olivares, A. L., Allison, S. D., and Treseder, K. K.: Soil microbes and their response to experimental warming over time: A meta-analysis of field studies, Soil Biol. Biochem., 107, 32–40, https://doi.org/10.1016/j.soilbio.2016.12.026, 2017.
Rousk, J. and Bååth, E.: Fungal biomass production and turnover in soil estimated using the acetate-in-ergosterol technique, Soil Biol. Biochem., 39, 2173–2177, https://doi.org/10.1016/j.soilbio.2007.03.023, 2007.
Rousk, J., Brookes, P. C., and Bååth, E.: Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization, Appl. Environ. Microbiol., 75, 1589–1596, https://doi.org/10.1128/AEM.02775-08, 2009.
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., Knight, R., and Fierer, N.: Soil bacterial and fungal communities across a pH gradient in an arable soil, ISME J., 4, 1340–1351, https://doi.org/10.1038/ismej.2010.58, 2010.
Sanderman, J., Hengl, T., and Fiske, G. J.: Soil carbon debt of 12,000 years of human land use, P. Natl. Acad. Sci. USA, 114, 9575–9580, https://doi.org/10.1073/pnas.1706103114, 2017.
Schimel, D., Stephens, B. B., and Fisher, J. B.: Effect of increasing CO2 on the terrestrial carbon cycle, P. Natl. Acad. Sci. USA, 112, 436–441, https://doi.org/10.1073/pnas.1407302112, 2015.
Sengupta, A., Fansler, S. J., Chu, R. K., Danczak, R. E., Garayburu-Caruso, V. A., Renteria, L., Song, H.-S., Toyoda, J., Wells, J., and Stegen, J. C.: Disturbance triggers non-linear microbe–environment feedbacks, Biogeosciences, 18, 4773–4789, https://doi.org/10.5194/bg-18-4773-2021, 2021.
Shi, Z., Crowell, S., Luo, Y., and Moore, B.: Model structures amplify uncertainty in predicted soil carbon responses to climate change, Nat. Commun., 9, 2171, https://doi.org/10.1038/s41467-018-04526-9, 2018.
Six, J., Frey, S. D., Thiet, R. K., and Batten, K. M.: Bacterial and Fungal Contributions to Carbon Sequestration in Agroecosystems, Soil Sci. Soc. Am. J., 70, 555, https://doi.org/10.2136/sssaj2004.0347, 2006.
Soares, M. and Rousk, J.: Microbial growth and carbon use efficiency in soil: Links to fungal-bacterial dominance, SOC-quality and stoichiometry, Soil Biol. Biochem., 131, 195–205, https://doi.org/10.1016/j.soilbio.2019.01.010, 2019.
Strickland, M. S. and Rousk, J.: Considering fungal: Bacterial dominance in soils – Methods, controls, and ecosystem implications, Soil Biol. Biochem., 42, 1385–1395, https://doi.org/10.1016/j.soilbio.2010.05.007, 2010.
Sulman, B. N., Phillips, R. P., Oishi, A. C., Shevliakova, E., and Pacala, S. W.: Microbe-driven turnover offsets mineral-mediated storage of soil carbon under elevated CO2, Nat. Clim. Chang., 4, 1385–1395, https://doi.org/10.1038/nclimate2436, 2014.
Tagesson, T., Schurgers, G., Horion, S., Ciais, P., Tian, F., Brandt, M., Ahlström, A., Wigneron, J. P., Ardö, J., Olin, S., Fan, L., Wu, Z., and Fensholt, R.: Recent divergence in the contributions of tropical and boreal forests to the terrestrial carbon sink, Nat. Ecol. Evol., 4, 202–209, https://doi.org/10.1038/s41559-019-1090-0, 2020.
Tedersoo, L., Bahram, M., Põlme, S., Kõljalg, U., Yorou, N. S., Wijesundera, R., Ruiz, L. V., Vasco-Palacios, A. M., Thu, P. Q., Suija, A., Smith, M. E., Sharp, C., Saluveer, E., Saitta, A., Rosas, M., Riit, T., Ratkowsky, D., Pritsch, K., Põldmaa, K., Piepenbring, M., Phosri, C., Peterson, M., Parts, K., Pärtel, K., Otsing, E., Nouhra, E., Njouonkou, A. L., Nilsson, R. H., Morgado, L. N., Mayor, J., May, T. W., Majuakim, L., Lodge, D. J., Lee, S., Larsson, K. H., Kohout, P., Hosaka, K., Hiiesalu, I., Henkel, T. W., Harend, H., Guo, L. D., Greslebin, A., Grelet, G., Geml, J., Gates, G., Dunstan, W., Dunk, C., Drenkhan, R., Dearnaley, J., De Kesel, A., Dang, T., Chen, X., Buegger, F., Brearley, F. Q., Bonito, G., Anslan, S., Abell, S., and Abarenkov, K.: Global diversity and geography of soil fungi, Science, 346, 1256688, https://doi.org/10.1126/science.1256688, 2014.
Terrer, C., Phillips, R. P., Hungate, B. A., Rosende, J., Pett-Ridge, J., Craig, M. E., van Groenigen, K. J., Keenan, T. F., Sulman, B. N., Stocker, B. D., Reich, P. B., Pellegrini, A. F. A., Pendall, E., Zhang, H., Evans, R. D., Carrillo, Y., Fisher, J. B., Van Sundert, K., Vicca, S., and Jackson, R. B.: A trade-off between plant and soil carbon storage under elevated CO2, Nature, 591, 599–603, https://doi.org/10.1038/s41586-021-03306-8, 2021.
van den Hoogen, J., Geisen, S., Routh, D., Ferris, H., Traunspurger, W., Wardle, D. A., de Goede, R. G. M., Adams, B. J., Ahmad, W., Andriuzzi, W. S., Bardgett, R. D., Bonkowski, M., Campos-Herrera, R., Cares, J. E., Caruso, T., de Brito Caixeta, L., Chen, X., Costa, S. R., Creamer, R., Mauro da Cunha Castro, J., Dam, M., Djigal, D., Escuer, M., Griffiths, B. S., Gutiérrez, C., Hohberg, K., Kalinkina, D., Kardol, P., Kergunteuil, A., Korthals, G., Krashevska, V., Kudrin, A. A., Li, Q., Liang, W., Magilton, M., Marais, M., Martín, J. A. R., Matveeva, E., Mayad, E. H., Mulder, C., Mullin, P., Neilson, R., Nguyen, T. A. D., Nielsen, U. N., Okada, H., Rius, J. E. P., Pan, K., Peneva, V., Pellissier, L., Carlos Pereira da Silva, J., Pitteloud, C., Powers, T. O., Powers, K., Quist, C. W., Rasmann, S., Moreno, S. S., Scheu, S., Setälä, H., Sushchuk, A., Tiunov, A. V., Trap, J., van der Putten, W., Vestergård, M., Villenave, C., Waeyenberge, L., Wall, D. H., Wilschut, R., Wright, D. G., Yang, J., and Crowther, T. W.: Soil nematode abundance and functional group composition at a global scale, Nature, 572, 194–198, https://doi.org/10.1038/s41586-019-1418-6, 2019.
Van Der Heijden, M. G. A., Bardgett, R. D., and Van Straalen, N. M.: The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems, Ecol. Lett., 11, 296–310, https://doi.org/10.1111/j.1461-0248.2007.01139.x, 2008.
Waldrop, M. P., Holloway, J. M., Smith, D. B., Goldhaber, M. B., Drenovsky, R. E., Scow, K. M., Dick, R., Howard, D., Wylie, B., and Grace, J. B.: The interacting roles of climate, soils, and plant production on soil microbial communities at a continental scale, Ecology, 98, 1957–1967, https://doi.org/10.1002/ecy.1883, 2017.
Wang, Q., Liu, S., and Tian, P.: Carbon quality and soil microbial property control the latitudinal pattern in temperature sensitivity of soil microbial respiration across Chinese forest ecosystems, Glob. Chang. Biol., 24, 2841–2849, https://doi.org/10.1111/gcb.14105, 2018.
Waring, B. G., Averill, C., and Hawkes, C. V.: Differences in fungal and bacterial physiology alter soil carbon and nitrogen cycling: Insights from meta-analysis and theoretical models, Ecol. Lett., 16, 887–894, https://doi.org/10.1111/ele.12125, 2013.
Wieder, W. R., Bonan, G. B., and Allison, S. D.: Global soil carbon projections are improved by modelling microbial processes, Nat. Clim. Chang., 3, 909–912, https://doi.org/10.1038/nclimate1951, 2013.
Wieder, W. R., Allison, S. D., Davidson, E. A., Georgiou, K., Hararuk, O., He, Y., Hopkins, F., Luo, Y., Smith, M. J., Sulman, B., Todd-Brown, K., Wang, Y. P., Xia, J., and Xu, X.: Explicitly representing soil microbial processes in Earth system models, Global Biogeochem. Cycles, 29, 1782–1800, https://doi.org/10.1002/2015GB005188, 2015.
Yu, K.: Biogeography-of-soil-microbes, figshare [data set], https://doi.org/10.6084/m9.figshare.19556419, 2022.
Yu, K. and D'Odorico, P.: Hydraulic lift as a determinant of tree-grass coexistence on savannas, New Phytol., 207, 1038–1051, https://doi.org/10.1111/nph.13431, 2015.
Yu, K., Smith, W. K., Trugman, A. T., Condit, R., Hubbell, S. P., Sardans, J., Peng, C., Zhu, K., Peñuelas, J., Cailleret, M., Levanic, T., Gessler, A., Schaub, M., Ferretti, M., and Anderegg, W. R. L.: Pervasive decreases in living vegetation carbon turnover time across forest climate zones, P. Natl. Acad. Sci. USA, 116, 24662–24667, https://doi.org/10.1073/pnas.1821387116, 2019.
Yue, H., Wang, M., Wang, S., Gilbert, J. A., Sun, X., Wu, L., Lin, Q., Hu, Y., Li, X., He, Z., Zhou, J., and Yang, Y.: The microbe-mediated mechanisms affecting topsoil carbon stock in Tibetan grasslands, ISME J., 9, 2012–2020, https://doi.org/10.1038/ismej.2015.19, 2015.
Zhu, Q., Riley, W. J., and Tang, J.: A new theory of plant-microbe nutrient competition resolves inconsistencies between observations and model predictions, Ecol. Appl., 27, 875–886, https://doi.org/10.1002/eap.1490, 2017.




