the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
TiP-Leaf: a dataset of leaf traits across vegetation types on the Tibetan Plateau
Yili Jin
Haoyan Wang
Jie Xia
Kai Li
Ying Hou
Jing Hu
Linfeng Wei
Kai Wu
Haojun Xia
Borui Zhou
Functional trait databases are emerging as a crucial tool for a wide range of ecological studies, including next-generation vegetation modelling across the world. However, few large-scale studies have been reported on plant traits in the Tibetan Plateau (TP), the cradle of East Asian flora and fauna with specific alpine ecosystems, and no report on plant trait databases could be found. In this work, an extensive dataset of 11 leaf functional traits (TiP-Leaf), mainly for herbs and shrubs and a few trees on the TP, was compiled through field surveys. The TiP-Leaf dataset, which was compiled from 336 sites distributed mainly on the plateau surface and the northern margin of the TP across alpine and temperate vegetation regions and sampled from 2018 to 2021, contained 1692 morphological trait measurements of leaf thickness, leaf fresh weight, leaf dry weight, leaf dry-matter content, leaf water content, leaf area, specific leaf area and leaf mass per area and 1645 chemical element trait measurements of leaf carbon, nitrogen and phosphorus contents. Thus, 468 species that belong to 184 genera and 51 families were obtained and measured. In addition to leaf trait measurements, the geographic coordinates, bioclimate variables, disturbance intensities and vegetation types of each site were also recorded. The dataset could provide solid data support to effectively quantify the modern ecological features of alpine ecosystems, thereby further evaluating the response of alpine ecosystems to climate change and human disturbances and improving the next-generation vegetation model. The dataset, which is available from the National Tibetan Plateau Data Center (TPDC; Jin et al., 2022a; https://doi.org/10.11888/Terre.tpdc.272516), can make a great contribution to the regional and global plant trait databases.
- Article
(2676 KB) - Full-text XML
-
Supplement
(621 KB) - BibTeX
- EndNote
Plant traits of morphological, anatomical, physiological and phenological characteristics respond to changes in the living environment, affect ecosystem functions (Díaz and Cabido, 2001) and drive species coexistence under environmental constraints (Violle et al., 2007). Over the past 3 decades, a growing body of trait analyses has quantified the global and regional distribution patterns of key functional traits, such as leaf (Reich and Oleksyn, 2004; Wright et al., 2004), seed size (Moles et al., 2007), plant height (Moles et al., 2009), wood (Chave et al., 2009), plant form and function (Díaz et al., 2016), root (Ma et al., 2018) and flower (Roddy et al., 2021). Such studies have successfully linked plant traits with environmental changes (Meng et al., 2009, 2015; Myers-Smith et al., 2019; Maes et al., 2020; Wang et al., 2022), natural and anthropogenic disturbances (Díaz et al., 2007) and ecosystem functions (Reichstein et al., 2014). Findings from plant trait–environment–ecosystem function interaction could be further utilised to map the spatial pattern of plant traits (Butler et al., 2018), build the next generation of vegetation models (Berzaghi et al., 2020), predict vegetation distribution (van Bodegom et al., 2014) and function (Wang et al., 2017) and be incorporated into the Earth system model (Wullschleger et al., 2014). New insights into ecosystem traits (He et al., 2019) and the trait network (He et al., 2020) are bridging multiple dimensions of biology, macroecology and geoscience. All these works require global and regional plant trait databases, such as the TRY (Kattge et al., 2011, 2020), Growth-Form (Taseski et al., 2019), Global Inventory of Floras and Traits (Weigelt et al., 2019), Fine-Root (Iversen et al., 2017), GRoot (Guerrero-Ramírez et al., 2021) and tundra traits (Bjorkman et al., 2018) and the Plant Trait for Mediterranean Basin Species (BROT) (Tavşanoğlu and Pausas, 2018), China traits (Wang et al., 2018), AusTraits (Falster et al., 2021) and LT-Brazil (Mariano et al., 2021).
Plant trait databases across various biomes at global, continental and regional scales have increased greatly, even in some remote areas with logistical difficulties, including the tundra (Bjorkman et al., 2018) and tropical (Mariano et al., 2021) regions. However, the Earth still has undersampled regions. The Tibetan Plateau (TP) includes the two major regions of Qinghai Province and the Xizang Autonomous Region and partial areas from north-western Gansu Province, southern Xinjiang Autonomous Region, western Sichuan Province and north-western Yunnan Province in China and is also called the “Qinghai–Tibetan Plateau”. As the world's “Third Pole”, the “Asia Water Tower” and the cradle of East Asian flora, the TP is the most underrepresented region in global and regional plant trait databases. The first version of the Chinese plant trait database (Wang et al., 2018) does not contain data from the TP, and the global plant trait database TRY (Kattge et al., 2020) has only a few collections from various sources with non-systematic sampling. Field-based, local studies of plant functional traits on the TP have made some interesting advances. For example, Luo et al. (2005) linked the plant traits with ecosystem functions, He et al. (2006) explored the influencing factors on plant traits, Geng et al. (2014) quantified the patterns of plant trait correlations between above- and below-ground components, Wang et al. (2020) compared their work with the global dataset, and Xu et al. (2021) analysed the mechanism of plant trait variation along the altitude pattern. However, such works, where the sampling sites have been mostly along the main roads in the eastern TP, were also limited. Plant trait records from the central to western TP are extremely rare. However, the TP has the richest temperate alpine flora (Ding et al., 2020) and the most abundant plant diversity in the world (Wang and Hong, 2022). It was also an evolutionary cradle for the herbaceous genera of China (Lu et al., 2018). The uplift of the TP and its unique alpine vegetation are important to the monsoon climate system and vegetation of East Asia (Chang, 1983) and regional and global climate change studies (Piao et al., 2019).
As the largest and highest plateau in the world, the TP has not only changed the regional and global climate system, geological structure, topography and hydrology (Yao et al., 2012), but has also influenced the evolution of the flora, fauna and biodiversity strongly (Ding et al., 2020). It has 8876 vascular species from 1371 genera and 211 families, including 6475 herbaceous and 2401 woody plants, of which 1706 were endemic to the TP (Yan et al., 2013), and has three biodiversity hotspots of the world (Sloan et al., 2014; Wang and Hong, 2022). Vegetation changes from south-east to north-west, from lowland broad-leaved evergreen forests, including tropical rainforest and subtropical evergreen forest, montane mixed evergreen and deciduous forest, subalpine conifer forest to alpine shrubland, meadow, steppe and desert along an annual precipitation gradient from ca. 3000 to 50 mm (Chang, 1983). In physiognomy, the unique alpine vegetation looks similar to the arctic tundra but has different species composition. The plateau has amplified changes in climates (Chen et al., 2015), and rapid climate change has led to profound changes in alpine species and ecosystems (Zhang et al., 2015; Piao et al., 2019). Plant traits, as the link amongst species, environment and ecosystem functions, are the best tools to study the impacts of climate change on vegetation. Therefore, the establishment of the TP plant trait database and further analysis of plant trait–environment–ecosystem function relationships are of great significance to understanding the future change and sustainable development of the unique alpine vegetation on the roof of the world.
In this work, a TP leaf trait dataset (TiP-Leaf) was established, and 11 leaf traits from 468 species of 1692 leaf samples were collected from 336 sites across five of the six vegetation types on the TP. The climate data of the sites were also provided. This dataset is not only an update of the Chinese plant trait database, but is also a great contribution to the global trait database.
The leaf traits of the dominant and common plant species of the TP distributed mostly across the plateau surface were sampled and measured from July to August in the summer of 2018–2021. Vegetation surveys were conducted in four regions (Fig. 1), namely the source area of the Three Rivers in Qinghai Province in the north-eastern TP (2018), the southern Xizang Autonomous Region in the south-eastern and central TP (2019), Ngari Prefecture in the north-western TP and the Qaidam Basin of the north-eastern TP (2020), and the Qilian Mountains, the Altun Mountains and the Kunlun Mountains in the northern margin of the TP, passing through the southern margin of the Tarim Basin in Xinjiang Autonomous Region (2021).
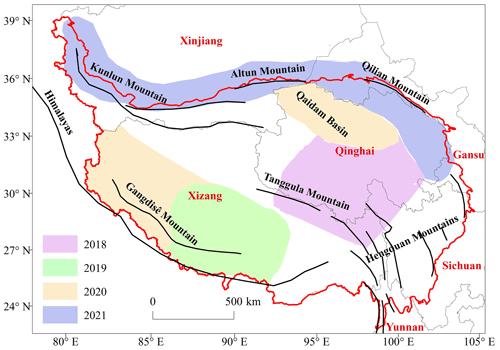
Figure 1Location and administrative division of the TP. The red line indicates the boundary of the TP in China, which involves six administrative divisions (light black lines): Xizang, Qinghai, Sichuan, Xinjiang, Gansu and Yunnan. The bold black lines represent important mountains. Four blocks with different colours represent the approximate areas of four investigations conducted in various years. The background map is from the Chinese National Bureau of Surveying and Mapping.
3.1 Sampling sites
Considering the zonal vegetation types and the precipitation gradient, 336 sites (Fig. 2) were selected to investigate the vegetation with less grazing and other anthropogenic disturbances. Shrubby and herbaceous vegetation was mainly selected (332 sites) along with the forest vegetation at the four sites (but removed for further analysis). At each site, one to three plots were set up to survey the species composition, abundance, coverage and plant height. The plot areas for herbaceous vegetation, shrubby vegetation and forest vegetation were 1 m × 1 m, 2 m × 2 m or 5 m × 5 m and 10 m × 10 m, respectively. Geographical locations, natural and human disturbances, and vegetation types were also recorded (Jin et al., 2022b). The dominant and common plant species at each site were determined by visual inspection, the leaf samples of these plants were picked up, and the leaf traits were measured. Root samples were obtained using the soil pit method. The root traits were also measured but are not shown in this paper.
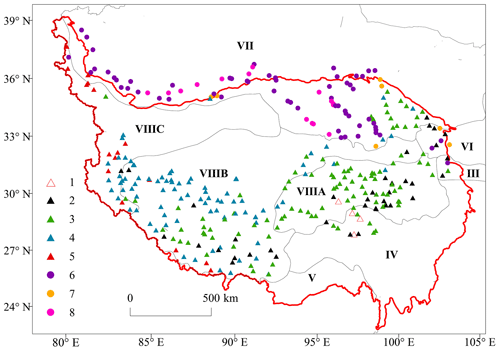
Figure 2Site distribution of the TiP-Leaf dataset. Vegetation regions were extracted from the vegetation regionalisation of China (ECVMC, 2007b): III, Warm Temperate Deciduous Broad-leaf Forest Region; IV, Subtropical Broad-leaf Evergreen Forest Region; V, Tropical Monsoon Rain Forest and Rain Forest Region; VI, Temperate Steppe Region; VII, Temperate Desert Region; VIII, TP Alpine Vegetation Region. VIIIA, East TP Alpine Scrub and Alpine Meadow Subregion; VIIIB, Middle TP Alpine Steppe Subregion; VIIIC, Northwest TP Alpine Desert Subregion. Vegetation types were classified on the basis of field records. Numbers indicate the vegetation types recorded in the field. 1, coniferous forest; 2, alpine shrubland; 3, alpine meadow; 4, alpine steppe; 5, alpine desert; 6, temperate desert; 7, temperate steppe; 8, temperate meadow. The background map is from the Chinese National Bureau of Surveying and Mapping.
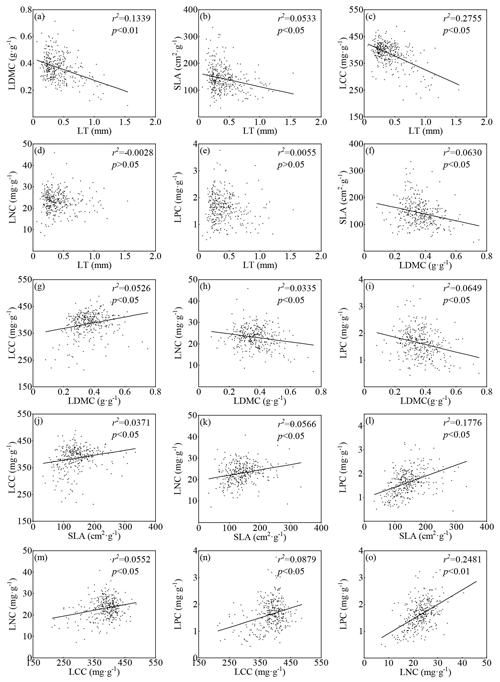
Figure 3Relationship between the site-based average of key leaf traits in the TiP-Leaf dataset. The black dot indicates the mean leaf trait measurements of all species at the site, and the straight line represents the fitting of the linear model. r2 is the adjusted r2, and p represents the probability value of the regression model.
The vegetation of the TP was classified into eight types: high-cold (alpine) shrubland, meadow, steppe and desert, temperate steppe, meadow and desert, and coniferous forest, on the basis of field records (Fig. 2). Alpine shrubland is dominated by evergreen broad-leaved shrubs (Rhododendron), deciduous broad-leaved shrubs (Salix, Dasiphora and Sibiraea) and evergreen coniferous shrubs (Juniperus) that are distributed in the cold and semi-humid south-eastern TP (ca. 600–1000 mm yr−1). Alpine meadow is widely developed in the eastern TP, where cold and wet climates are prevalent (ca. 600 mm yr−1), dominated by several Kobresia species and mixed in with perennial forbs and cushion plants. Alpine steppe is in the central TP, with a large continuous distribution adapted to the cold and semi-dry continental climate (ca. 200 mm yr−1) and mainly composed of Stipa and Artemisia. Alpine desert is mainly distributed in the north-western TP, where the climate is extremely continental (ca. 50 mm yr−1) and dominated by Krascheninnikovia compacta and Ajania tibetica. Temperate meadow, steppe and desert are distributed on the northern margin of the TP and the Qaidam Basin of the north-eastern TP, where elevations are lower and the climate is relatively dry, dominated by several xerophytes, especially Haloxylon ammodendron, Halogeton glomeratus, Phragmites australis, Ephedra, Kalidium, Calligonum and Tamarix. Subalpine coniferous (Abies and Picea) forests are found on the south-eastern and eastern margins. Therefore, the vegetation of the plateau is distributed along a transitional gradient from south-east to north-west, ranging from subalpine forests, alpine meadow and scrub to alpine steppe and temperate desert to alpine desert. The alpine vegetation in the vegetation classification of China was usually called high-cold vegetation (ECVMC, 2007a). Lowland tropical and montane subtropical evergreen forests do not exist in the sampling area. Hence, they are not included in this study.
Each site was also assigned to a vegetation region on the basis of the vegetation regionalisation of China (ECVMC, 2007b). The TP has six vegetation regions, namely the Alpine Vegetation Region, Temperate Steppe Region, Temperate Desert Region, Warm Temperate Deciduous Broad-leaf Forest Region, Subtropical Broad-leaf Evergreen Forest Region and Tropical Monsoon Rain Forest and Rain Forest Region. The sampling sites were mainly concentrated in the Alpine Vegetation Region. Therefore, in accordance with the degree of drought, the Selianinov drought index used in the Vegetation Regionalisation Map of China (ECVMC, 2007b), TP vegetation was further divided into three subregions from south-east to north-west, namely the East TP Alpine Scrub and Alpine Meadow Subregion, the Middle TP Alpine Steppe Subregion and the Northwest TP Alpine Desert Subregion.
The plant name was determined in accordance with Flora Reipublicae Popularis Sinicae (Editorial Committee of Flora of China, 1959–2004), Flora Qinghaiica (Editorial Committee of the Flora Qinghaiica, 1996–1999), Flora Xizangica (Integrated Scientific Expedition to Qinghai-Tibet Plateau, Chinese Academy of Sciences, 1983–1987), Flora of Gansu (Editorial Committee of Flora of Gansu, 2005), Flora Xinjiangensis (Commissione Redactorum Flora Xinjiangensis, 1992–1996) and Flora in Desertis Reipublicae Populorum Sinarum (Liu, 1985–1992). The final species correction was based on the iPlant website (http://www.iplant.cn/, last access: 7 April 2022), which merged all of the information from the Chinese and English versions of Flora of China on the basis of the APG IV classification (Angiosperm Phylogeny Group, 2016).
3.2 Leaf trait measurements
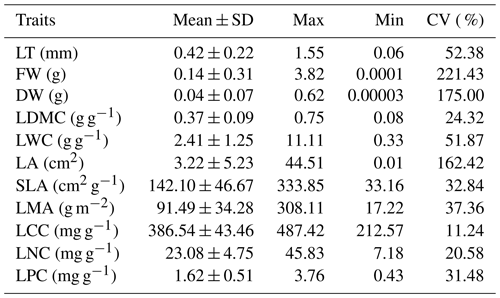
At each site, two to three mature and disease-free complete leaves from each individual of dominant and common plant species were collected, and at least 30 individuals were selected to meet the needs of trait measurement and element analysis. When the single leaf was small, microphyllous or leptophyllous, 100–200 leaves were picked. In total, 11 leaf functional traits (e.g. leaf thickness, LT; fresh weight, FW; dry weight, DW; leaf dry-matter content, LDMC; leaf water content, LWC; leaf area, LA; specific leaf area, SLA; leaf mass per area, LMA; leaf carbon content, LCC; leaf nitrogen content, LNC; leaf phosphorus content, LPC) were measured and calculated on the basis of the handbook of standardised measurement for plant functional traits worldwide (Cornelissen et al., 2003; Pérez-Harguindeguy et al., 2013).
LT (mm) was measured on the sampling day by using Vernier callipers with an accuracy of 0.01 mm. The thickness in the middle of the vein and margin of each leaf was measured, and then the average of the five leaves was taken as the LT of a species. In addition to LT, 20–30 leaves for normal-leaved plants and 100–200 leaves for small- to leptophyll-leaved plants were generally selected for other trait variable measurements. FW (g) was obtained by weighing with a electronic balance. Subsequently, the fresh leaves were oven dried at 75 ∘C for 48–72 h to obtain the DW (g). LDMC was measured as follows: LDMC (g g−1) = DWFW. LA (cm2) was measured using a scanner (EPSON Perfection V 700 Photo Scanner) and software (WinFOLIA Pro, Canada). SLA and LMA were measured as follows: SLA (cm2 g−1) = LADW and LMA (g m−2) = DWLA × 104. The dried leaves were further used for chemical analysis. LCC (mg g−1), LNC (mg g−1) and LPC (mg g−1) were determined by the outside-temperature hot potassium dichromate oxidation–volumetric method (Wu, 2007), the distillation–titration method and the vanadium molybdate yellow colorimetric method (Fang et al., 2011), respectively.
3.3 Climate data
The climate data of each sampling site were extracted from the climate and bioclimate datasets of China (Cheng et al., 2022; Wei et al., 2022). China's climate dataset consists of three variables (monthly temperature, precipitation and sunshine percentage) that were averaged from long-term records from 1981 to 2010 at 2152 meteorological stations across China (China Meteorological Data Service Centre, http://data.cma.cn, last access: 1 June 2021). These three climate factors and the absolute maximum and minimum temperatures during the 30-year period of 1981–2010 were interpolated into 1 km grid cells by using a surface fitting technique of a thin-plate smoothing spline (ANUSPLIN version 4.4, Hutchinson and Xu, 2013; Xu and Hutchinson, 2013) that considered the impact of elevation on climates on the basis of the digital elevation model of the Shuttle Radar Topography Mission (Farr et al., 2007). The interpolated climate data were used to drive a bioclimate software (Gallego-Sala et al., 2010) to calculate the mean annual temperature (MAT), mean temperature of the coldest month (MTCO), mean temperature of the warmest month (MTWA), annual growing degree days above 0 (GDD0) and 5 ∘C (GDD5), mean annual precipitation (MAP), growing season precipitation (GP), annual drought index (1-AET/PET), and annual moisture index (MAP/PET), where AET and PET refer to the annual actual evapotranspiration and annual potential evapotranspiration, respectively.
3.4 Data analysis
Besides the data description of leaf trait characteristics, six key leaf functional traits (e.g. LT, LDMC, SLA, LCC, LNC and LPC) that reflect the key ecological significance of plants that grew at high altitude and in an extremely cold environment were selected in this paper for further simple statistical analyses. LT affects the water supply and storage of leaves and the exchange process of matter and energy in photosynthesis. LDMC reflects the ability of plants to acquire surrounding environmental resources. SLA is considered the first-choice index for studying plant physiological and ecological strategies under specific environmental conditions. LCC is the main structural material of plants. LNC characterises the ability of plants to absorb and utilise nutrient elements. LPC is the second-largest element that affects plant growth. The mean, minimum, maximum, standard deviation and coefficient of variation (CV) of traits at each site were calculated to generally show the pattern of leaf traits of the Tibetan ecosystems. The linear relationships between leaf traits of the site average were analysed and mapped using the Origin software to reveal the trade-off between different traits in the special alpine ecosystem. The detailed analyses of all the leaf traits, their variations and their spatial patterns within and amongst functional groups and at species and site levels will be further analysed in another paper.
4.1 Spatial distribution of sites
A total of 11 key plant leaf traits of 1692 individuals of 468 species from 336 sites were measured (Figs. 1 and 2).
The sampling sites were located on the north-eastern, central to south-western and northern margins of the TP, along with 145 sites in Xizang, 121 sites in Qinghai, 43 sites in Xinjiang, 16 sites in Gansu and 11 sites in Sichuan (Fig. 1). The south-eastern TP, where forest ecosystems are distributed, has few plant trait data. However, field measurements are being conducted in the Hengduan Mountains to measure the leaf, twig and root traits of dominant and common trees and shrubs. Other ecologists have worked on some parts of this region to perform leaf and other trait studies (Luo et al., 2005; Shi et al., 2012; Vandvik et al., 2020; Xu et al., 2021). The Hoh Xil dead zone in the central northern to north-western TP is logistically not accessible during the plant growing season when the frozen ground is melting. Therefore, the plant trait data have been unavailable to date.
4.2 Altitudinal range of sites
The altitudinal range of the sampling sites was between 805 and 5343 m, in which 69.3 % of the sites were located at the high altitudes (>3500 m), 18.5 % of the sites were located in the Qaidam Basin and eastern Qinghai with lower altitudes (2500–3500 m) and 12.2 % of the sites were located on the northern margin of the TP with the lowest altitudes (<2500 m).
4.3 Vegetation types of sites
In accordance with the field records, the vegetation was divided into eight types, along with 108 sites in alpine meadow, 87 sites in alpine steppe, 61 sites in temperate desert, 38 sites in alpine shrubland, 16 sites in alpine desert, 15 sites in temperate meadow, 7 sites in temperate steppe and 4 sites in forest. In addition, the number of sites in the TP Alpine Vegetation Region was the most abundant (63.1 %), including its three subregions, namely the Middle TP Alpine Steppe Subregion (33.9 %), the East TP Alpine Scrub and Alpine Meadow Subregion (20.8 %) and the Northwest TP Alpine Desert Subregion (8.3 %), followed by the Temperate Desert Region (29.4 %) and other vegetation regions (7.5 %), which are the Subtropical Evergreen Broad-leaved Forest Region (3.9 %), the Temperate Steppe Region (2.4 %) and the Warm Temperate Deciduous Broad-leaved Forest Region (1.2 %), as shown in Fig. 2.
5.1 Plant species
A total of 1692 leaf samples were collected and measured in the TiP-Leaf dataset, including 468 species that belong to 184 genera in 51 families (amongst them, 17 samples were identified as genera, 6 samples were identified as families and 1 sample could not be identified). Some species were sampled frequently. For example, Kobresia pygmaea occurred 52 times, mainly in the eastern and southern TP. Stipa purpurea occurred 47 times, mainly in the central and western TP. Potentilla bifurca occurred 41 times, mainly in the southern, western and north-eastern TP. However, at some sites, only one or two species were sampled, especially in the Qaidam Basin and the northern margin of the TP. The top five families with the largest number of sampled species were as follows: Asteraceae (83 species and 24 genera), Poaceae (47 species and 18 genera), Fabaceae (46 species and 11 genera), Cyperaceae (29 species and 4 genera) and Rosaceae (28 species and 10 genera). Amongst the 468 species, 79 species, including Rhodiola smithii, Pomatosace filicula, Oxytropis sericopetala, Arenaria gerzensis, Onosma waltonii, Delphinium qinghaiense, Metaeritrichium microuloides and Androsace cuttingii, were unique to the TP. Furthermore, two (Rosa rugosa and Rheum globulosum) were endangered species, seven (Arnebia guttata, Tamarix taklamakanensis, Rhodiola smithii, Juniperus tibetica, Reaumuria kaschgarica, Rheum tanguticum and Metaeritrichium microuloides) were vulnerable species and 10 (Myricaria prostrata, Euphorbia kozlovii, Hippophae tibetana, Phlomis pygmaea, Physochlaina praealta, Gentiana siphonantha, Astragalus handelii, Androsace cuttingii, Carex nakaoana and Leiospora exscapa) were near-threatened species in the TiP-Leaf dataset.
5.2 Leaf trait variations
The site-level leaf traits are shown in Table 1. The variation of each leaf trait was significant. In particular, DW, FW and LA varied by more than 150 %, followed by LT and LWC. The variations of LMA, SLA, LDMC, LCC, LNC and LPC were slightly modest, relatively.
5.3 Leaf trait relationships
The fitting of linear models of the site averages of the six leaf traits (Fig. 3) showed that LT was significantly negatively correlated with LDMC (r2=0.1339; p<0.05; Fig. 3a), SLA (r2=0.0533; p<0.05; Fig. 3b) and LCC (r2=0.2755; p<0.05; Fig. 3c) with downward trends. No relationship was found between LT and LNC (; p>0.05; Fig. 3d) nor LPC (r2=0.0055; p>0.05; Fig. 3e). The results also revealed that LDMC was significantly negatively correlated with SLA (r2=0.0630; p<0.05; Fig. 3f), LNC (r2=0.0335; p<0.05; Fig. 3h) and LPC (r2=0.0649; p<0.05; Fig. 3i) and significantly positively correlated with LCC (r2=0.0526; p<0.05; Fig. 3g). In addition, linearly inversed relationships were observed between SLA and LCC (r2=0.0371; p<0.05; Fig. 3j) and LNC (r2=0.0566; p<0.05; Fig. 3k) and LPC (r2=0.1776; p<0.05; Fig. 3l). The three leaf chemical traits were also related to one another, and the relationship between LNC and LPC (r2=0.2481; p<0.05; Fig. 3o) was closer than that between LNC and LCC (r2=0.0552; p<0.05; Fig. 3m) and between LPC and LCC (r2=0.0879; p<0.05; Fig. 3n).
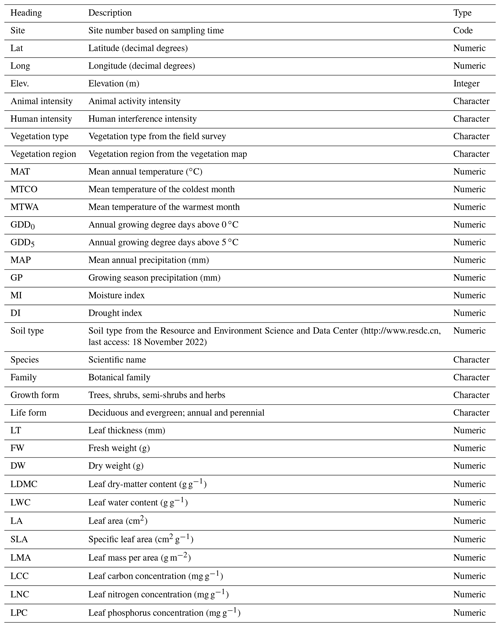
The TiP-Leaf dataset comprises three data sheets in Microsoft Excel format, namely (a) a data sheet named “variables”, which describes the header information of the geographical coordinates, climate and traits in the dataset (Table 2), (b) a data sheet (site information) that reports the site location and climate data and (c) a data sheet (plant traits) of the complete trait data of each plant species at each sampling site. As studies based on the TiP-Leaf dataset are already underway, researchers interested in using such data previously are strongly recommended to contact the authors to avoid overlapping studies. The dataset will be available through the National Tibetan Plateau Data Center (TPDC; Jin et al., 2022a; https://doi.org/10.11888/Terre.tpdc.272516) and shall also be made available via the global TRY plant trait database (Kattge et al., 2011, 2020; https://www.try-db.org/, last access: 18 July 2022).
The TiP-Leaf dataset was compiled from direct field measurements, covering a great proportion of plant species and vegetation types on the highest plateau in the world. The dataset provides an important data foundation not only for quantitative analyses of modern alpine vegetation, but also for the prediction of future responses of alpine ecosystems to climate change and improvement of next-generation vegetation models. It could also be used to promote vegetation protection and restoration on the TP and contribute to the global plant trait database. However, the dataset also presents some unavoidable limitations. For example, the establishment of sampling sites and the judgment of dominant and common species are mostly subjective. The leaves of some plants are extremely small, resulting in incomplete recognition when scanning the LA. Due to harsh field conditions, measuring the plant traits in time occasionally becomes impossible. Preventing some leaves from losing too much water to withering is still inevitable, although we have taken protective measures for the leaves. Inadequate collection of some leaf samples results in fewer data of plant chemical element content than that of morphological traits. In any case, performing large-scale collection of plant traits on the TP, which requires a lot of manpower and material resources as well as overcoming the adverse environment of high altitude and extreme variability, is not easy.
The dataset in this study provides more leaf trait measurements and covers more sampling sites, which were located not only along the main roads, but also the accessible pathlets, than previous studies (Luo et al., 2005; He et al., 2006; He et al., 2010; Geng et al., 2014; Wang et al., 2020; Xu et al., 2021). This dataset is the first plant trait one that represents all of the alpine vegetation on the TP. However, more collections of trait data are needed in remote areas with assessable difficulty, such as the Hoh Xil dead zone in the north-western TP (alpine meadow, steppe and desert vegetation) and the mountainous areas of the eastern and south-eastern TP with fewer trait studies (subalpine and alpine forest and shrubland vegetation). These works could enhance the representativeness of the whole TiP-Leaf trait database in terms of geographical space and vegetation type. Given the complex topography of the plateau, more sites are requested to be surveyed. Given the flourishing of alpine flora, traits from more plant species should be measured. At present, the TiP-Leaf dataset consists of leaf traits only. The TiP-Root trait dataset is underway, and the trait data of the twigs and branches of woody species will be measured further.
The supplement related to this article is available online at: https://doi.org/10.5194/essd-15-25-2023-supplement.
JN conceived the study. KL, YJ and JN led the field works. YJ, HW, JX, YH, JH, LW, KW, HX and BZ collected leaf samples and measured plant traits. YJ, HW and JX processed the dataset, performed the analyses and wrote the first draft. JN and YJ improved the manuscript. All the authors approved the final version of the submitted manuscript.
The contact author has declared that none of the authors has any competing interests.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the special issue “Extreme environment datasets for the three poles”. It is not associated with a conference.
The authors sincerely thank Chenyu Li, Tudan Luosang, Yezi Sheng, Pingyu Sun and Deyu Xu for their help in the field survey and Jun Li, Ang Liu, Rui Tang and Xinxin Zhou for helping with specimen identification.
This work was supported by the Second Tibetan Plateau Scientific Expedition and Research Program (STEP (grant no. 2019QZKK0402)) and the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDA2009000003).
This paper was edited by David Carlson and reviewed by two anonymous referees.
Angiosperm Phylogeny Group: An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV, Bot. J. Linn. Soc., 181, 1–20, https://doi.org/10.1111/boj.12385, 2016.
Berzaghi, F., Wright, I. J., Kramer, K., Oddou-Muratorio, S., Bohn, F. J., Reyer, C. P. O., Sabate, S., Sanders, T. G. M., and Hartig, F.: Towards a New Generation of Trait-Flexible Vegetation Models, Trends Ecol. Evol., 35, 191–205, https://doi.org/10.1016/j.tree.2019.11.006, 2020.
Bjorkman, A. D., Myers-Smith, I. H., Elmendorf, S. C., Normand, S., Thomas, H. J. D., Alatalo, J. M., Alexander, H., Anadon-Rosell, A., Angers-Blondin, S., Bai, Y., Baruah, G., te Beest, M., Berner, L., Bjork, R. G., Blok, D., Bruelheide, H., Buchwal, A ., Buras, A., Carbognani, M., Christie, K., Collier, L. S., Cooper, E. J., Cornelissen, J. H. C., Dickinson, K. J. M., Dullinger, S., Elberling, B., Eskelinen, A., Forbes, B. C., Frei, E. R., Iturrate-Garcia, M., Good, M. K., Grau, O., Green, P., Greve, M., Grogan, P., Haider, S., Hajek, T., Hallinger, M., Happonen, K., Harper, K. A., Heijmans, M. M. P. D., Henry, G. H. R., Hermanutz, L., Hewitt, R. E., Hollister, R. D., Hudson, J., Hulber, K., Iversen, C. M., Jaroszynska, F., Jimenez-Alfaro, B., Johnstone, J., Jorgensen, R. H., Kaarlejarvi, E., Klady, R., Klimesova, J., Korsten, A., Kuleza, S., Kulonen, A., Lamarque, L. J., Lantz, T., Lavalle, A., Lembrechts, J. J., Levesque, E., Little, C. J., Luoto, M., Macek, P., Mack, M. C., Mathakutha, R., Michelsen, A., Milbau, A., Molau, U., Morgan, J. W., Morsdorf, M. A., Nabe-Nielsen, J., Nielsen, S. S., Ninot, J. M., Oberbauer, S. F., Olofsson, J., Onipchenko, V. G., Petraglia, A., Pickering, C., Prevey, J. S., Rixen, C., Rumpf, S. B., Schaepman-Strub, G., Semenchuk, P., Shetti, R., Soudzilovskaia, N. A., Spasojevic, M. J., Speed, J. D. M., Street, L. E., Suding, K., Tape, K. D., Tomaselli, M., Trant, A., Treier, U. A., Tremblay, J. P., Tremblay, M., Venn, S., Virkkala, A. M., Vowles, T., Weijers, S., Wilmking, M., Wipf, S., and Zamin, T.: Tundra Trait Team: A database of plant traits spanning the tundra biome, Global Ecol. Biogeogr., 27, 1402–1411, https://doi.org/10.1111/geb.12821, 2018.
Butler, E. E., Datta, A., Flores-Moreno, H., Chen, M., Wythers, K. R., Fazayeli, F., Banerjee, A., Atkin, O. K., Kattge, J., Amiaud, B., Blonder, B., Boenisch, G., Bond-Lamberty, B., Brown, K. A., Byun, C., Campetella, G., Cerabolini, B. E. L., Cornelissen, J. H. C., Craine, J. M., Craven, D., de Vries, F. T., Diaz, S., Domingues, T. F., Forey, E., Gonzalez-Melo, A., Gross, N., Han, W. X., Hattingh, W. N., Hickler, T., Jansen, S., Kramer, K., Kraft, N. J. B., Kurokawa, H., Laughlin, D. C., Meir, P., Minden, V., Niinemets, U., Onoda, Y., Penuelas, J., Read, Q., Sack, L., Schamp, B., Soudzilovskaia, N. A., Spasojevic, M. J., Sosinski, E., Thornton, P. E., Valladares, F., van Bodegom, P. M., Williams, M., Wirth, C., and Reich, P. B.: Mapping local and global variability in plant trait distributions, P. Natl. Acad. Sci. USA, 114, E10937–E10946, https://doi.org/10.1073/pnas.1708984114, 2018.
Chang, D. H. S.: The Tibetan Plateau in relation to the vegetation of China, Ann. Mo. Bot. Gard., 70, 564–570, https://doi.org/10.2307/2992087, 1983.
Chave, J., Coomes, D., Jansen, S., Lewis, S. L., Swenson, N. G., and Zanne, A. E.: Towards a worldwide wood economics spectrum, Ecol. Lett., 12, 351–366, https://doi.org/10.1111/j.1461-0248.2009.01285.x, 2009.
Chen, D. L., Xu, B. Q., Yao, T. D., Guo, Z. T., Cui, P., Chen, F. H., Zhang, R. H., Zhang, X. Z., Zhang, Y. L., Fan, J., Hou, Z. Q., and Zhang, T. H.: Assessment of past, present and future environmental changes on the Tibetan Plateau, Chin. Sci. Bull., 60, 3025–3035, https://doi.org/10.1360/N972014-01370, 2015.
Cheng, Q., Wu, X. Q., Wei, L. F., Hu, X. F., and Ni, J.: 30-year average monthly/1-km climate variables dataset of China (1951–1980, 1981–2010), Digital J. Global Change Data Repository [data set], https://doi.org/10.3974/geodb.2022.06.03.V1, 2022.
Commissione Redactorum Flora Xinjiangensis: Flora Xinjiangensis, 6 volumes, Xinjiang Science & Technology & Hygiene Publishing House, Ürümqi, 1992–1996.
Cornelissen, J. H. C., Lavorel, S., Garnier, E., Díaz, S., Buchmann, N., Gurvich, D. E., Reich, P. B., ter Steege, H., Morgan, H. D., van der Heijden, M. G. A., Pausas, J. G., and Poorter, H.: A handbook of protocols for standardised and easy measurement of plant functional traits worldwide, Aust. J. Bot., 51, 335–380, https://doi.org/10.1071/BT02124, 2003.
Díaz, S. and Cabido, M.: Vive la différence: plant functional diversity matters to ecosystem processes, Trends Ecol. Evol., 16, 646–655, https://doi.org/10.1016/S0169-5347(01)02283-2, 2001.
Díaz, S., Lavorel, S., McIntyre, S., Falczuk, V., Casanoves, F., Milchunas, D. G., Skarpe, C., Rusch, G., Sternberg, M., Noy-Meir, I., Landsberg, J., Zhang, W., Clark, H., and Campbell, B. D.: Plant trait responses to grazing – a global synthesis, Glob. Change Biol., 13, 313–341, https://doi.org/10.1111/j.1365-2486.2006.01288.x, 2007.
Díaz, S., Kattge, J., Cornelissen, J. H. C., Wright, I. J., Lavorel, S., Dray, S., Reu, B., Kleyer, M., Wirth, C., Prentice, I. C., Garnier, E., Bonisch, G., Westoby, M., Poorter, H., Reich, P. B., Moles, A. T., Dickie, J., Gillison, A. N., Zanne, A. E., Chave, J., Wright, S. J., Sheremet'ev, S. N., Jactel, H., Baraloto, C., Cerabolini, B., Pierce, S., Shipley, B., Kirkup, D., Casanoves, F., Joswig, J. S., Gunther, A., Falczuk, V., Ruger, N., Mahecha, M. D., and Gorne, L. D.: The global spectrum of plant form and function, Nature, 529, 167–171, https://doi.org/10.1038/nature16489, 2016.
Ding, W. N., Ree, R. H., Spicer, R. A., and Xing, Y. W.: Ancient orogenic and monsoon-driven assembly of the world's richest temperate alpine flora, Science, 369, 578–581, https://doi.org/10.1126/science.abb4484, 2020.
Editorial Committee of the Flora of China: Flora Reipublicae Popularis Sinicae, 80 volumes, Science Press, Beijing, 1959–2004.
Editorial Committee of the Flora of Gansu: Flora of Gansu, Volume 2, Gansu Science and Technology Press, Lanzhou, ISBN 9787542410075, 2005.
Editorial Committee of the Flora Qinghaiica: Flora Qinghaiica, 4 volumes, Qinghai People’s Publishing House, Xining, 1996–1999.
Editorial Committee of Vegetation Map of China, the Chinese Academy of Sciences (ECVMC): Vegetation of China and its Geographical Pattern – Illustration of the Vegetation Map of the People's Republic of China (1:1 000 000), Geology Press, Beijing, ISBN 9787116051461, 2007a.
Editorial Committee of Vegetation Map of China, the Chinese Academy of Sciences (ECVMC): Vegetation Map of the People's Republic of China (1:1 000 000), Geology Press, Beijing, ISBN 9787116045132, 2007b.
Falster, D., Gallagher, R., Wenk, E. H., Wright, I. J., Indiarto, D., Andrew, S. C., Baxter, C., Lawson, J., Allen, S., Fuchs, A., Monro, A., Kar, F., Adams, M. A., Ahrens, C. W., Alfonzetti, M., Angevin, T., Apgaua, D. M. G., Arndt, S., Atkin, O. K., Atkinson, J., Auld, T., Baker, A., von Balthazar, M., Bean, A., Blackman, C. J., Bloomfeld, K., Bowman, D. M. J. S., Bragg, J., Brodribb, T. J., Buckton, G., Burrows, G., Caldwell, E., Camac, J., Carpenter, R., Catford, J. A, Cawthray, G. R., Cernusak, L. A., Chandler, G., Chapman, A. R., Cheal, D., Cheesman, A. W., Chen, S. C., Choat, B., Clinton, B., Clode, P. L., Coleman, H., Cornwell, W. K., Cosgrove, M., Crisp, M., Cross, E., Crous, K. Y., Cunningham, S., Curran, T., Curtis, E., Daws, M. I., DeGabriel, J. L., Denton, M. D., Dong, N., Du, P. Z., Duan, H. L., Duncan, D. H., Duncan, R. P., Duretto, M., Dwyer, J. M., Edwards, C., Esperon-Rodriguez, M., Evans, J. R., Everingham, S. E., Farrell, C., Firn, J., Fonseca, C. R., French, B. J, Frood, D., Funk, J. L., Geange, S. R., Ghannoum, O., Gleason, S. M., Gosper, C. R., Gray, E., Groom, P. K., Grootemaat, S., Gross, C., Guerin, G., Guja, L., Hahs, A. K., Harrison, M. T., Hayes, P. E., Henery, M., Hochuli, D., Howell, J., Huang, G., Hughes, L., Huisman, J., Ilic, J., Jagdish, A., Jin, D., Jordan, G., Jurado, E., Kanowski, J., Kasel, S., Kellermann, J., Kenny, B., Kohout, M., Kooyman, R. M., Kotowska, M. M., Lai, H. R., Laliberte, E., Lambers, H., Lamont, B. B., Lanfear, R., van Langevelde, F., Laughlin, D. C., Laugier-kitchener, B. A., Laurance, S., Lehmann, C. E. R., Leigh, A., Leishman, M. R., Lenz, T., Lepschi, B., Lewis, J. D., Lim, F., Liu, U., Lord, J., Lusk, C. H., Macinnis-Ng, C., McPherson, H., Magallon, S., Manea, A., Lopez-Martinez, A., Mayfield, M., McCarthy, J. K., Meers, T., van der Merwe, M., Metcalfe, D. J., Milberg, P., Mokany, K., Moles, A. T., Moore, B. D., Moore, N., Morgan, J. W., Morris, W., Muir, A., Munroe, S., Nicholson, A., Nicolle, D., Nicotra, A. B., Niinemets, U., North, T., O'Reilly-Nugent, A., O'Sullivan, O. S., Oberle, B., Onoda, Y., Ooi, M. K. J., Osborne, C. P., Paczkowska, G., Pekin, B., Pereira, C. G., Pickering, C., Pickup, M., Pollock, L. J., Poot, P., Powell, J. R., Power, S. A, Prentice, I. C., Prior, L., Prober, S. M., Read, J., Reynolds, V., Richards, A. E., Richardson, B., Roderick, M. L., Rosell, J. A., Rossetto, M., Rye, B., Rymer, P. D., Sams, M., Sanson, G., Sauquet, H., Schmidt, S., Schonenberger, J., Schulze, E. D., Sendall, K., Sinclair, S., Smith, B., Smith, R., Soper, F., Sparrow, B., Standish, R. J., Staples, T. L., Stephens, R., Szota, C., Taseski, G., Tasker, E., Thomas, F., Tissue, D. T., Tjoelker, M. G., Tng, D. Y. P., de Tombeur, F., Tomlinson, K., Turner, N. C., Veneklaas, E. J., Venn, S., Vesk, P., Vlasveld, C., Vorontsova, M. S., Warren, C. A., Warwick, N., Weerasinghe, L. K., Wells, J., Westoby, M., White, M., Williams, N. S. G., Wills, J., Wilson, P. G., Yates, C., Zanne, A. E., Zemunik, G., and Zieminska, K.: AusTraits, a curated plant trait database for the Australian flora, Sci. Data, 8, 254, https://doi.org/10.1038/s41597-021-01006-6, 2021.
Fang, J. B., Pang, R. L., Guo, L. L., Xie, H. Z., Li, J., Luo, J., Yu, H., Liu, Y., and Wu, F. K.: Determination of nitrogen, phosphorus and potassium in plants, China Agriculture Press, Beijing, NY/T 2017–2011, 2011.
Farr, T. G., Rosen, P. A., Caro, E., Crippen, R., Duren, R., Hensley, S., Kobrick, M., Paller, M., Rodriguez, E., Roth, L., Seal, D., Shaffer, S., Shimada, J., Umland, J., Werner M., Oskin, M., Burbank, D., and Alsdorf, D.: The shuttle radar topography mission, Rev. Geophys., 45, RG2004, https://doi.org/10.1029/2005RG000183, 2007.
Gallego-Sala, A. V., Clark, J. M., House, J. I., Orr, H. G., Prentice, I. C., Smith, P., Farewell, T., and Chapman, S. J.: Bioclimatic envelope model of climate change impacts on blanket peatland distribution in Great Britain, Clim. Res., 45, 151–162, https://doi.org/10.3354/cr00911, 2010.
Geng, Y., Wang, L., Jin, D. M., Liu, H. Y., and He, J. S.: Alpine climate alters the relationships between leaf and root morphological traits but not chemical traits, Oecologia, 175, 445–455, https://doi.org/10.1007/s00442-014-2919-5, 2014.
Guerrero-Ramírez, N. R., Mommer, L., Freschet, G. T., Iversen, C. M., McCormack, M. L., Kattge, J., Poorter, H., van der Plas, F., Bergmann, J., Kuyper, T. W., York, L. M., Bruelheide, H., Laughlin, D. C., Meier, I. C., Roumet, C., Semchenko, M., Sweeney, C. J., van Ruijven, J., Valverde-Barrantes, O. J., Aubin, I., Catford, J. A., Manning, P., Martin, A., Milla, R., Minden, V., Pausas, J. G., Smith, S. W., Soudzilovskaia, N. A., Ammer, C., Butterfield, B., Craine, J., Cornelissen, J. H. C., de Vries, F. T., Isaac, M. E., Kramer, K., Konig, C., Lamb, E. G., Onipchenko, V. G., Penuelas, J., Reich, P. B., Rillig, M. C., Sack, L., Shipley, B., Tedersoo, L., Valladares, F., van Bodegom, P., Weigelt, P., Wright, J. P., and Weigelt, A.: Global root traits (GRooT) database, Global Ecol. Biogeogr., 30, 25–37, https://doi.org/10.1111/geb.13179, 2021.
He, J. S., Wang, Z. H., Wang, X. P., Schmid, B., Zuo, W. Y., Zhou, M., Zheng, C. Y., Wang, M. F., and Fang, J. Y.: A test of the generality of leaf trait relationships on the Tibetan Plateau, New Phytol., 170, 835–848, https://doi.org/10.1111/j.1469-8137.2006.01704.x, 2006.
He, J. S., Wang, X. P., Schmid, B., Flynn, D. F. B., Li, X. F., Reich, P. B., and Fang, J. Y.: Taxonomic identity, phylogeny, climate and soil fertility as driver of leaf traits across Chinese grassland biomes, J. Plant. Res., 123, 551–561, https://doi.org/10.1007/s10265-009-0294-9, 2010.
He, N. P., Liu, C. C., Piao, S. L., Sack, L., Xu, L., Luo, Y. Q., He, J. S., Han, X. G., Zhou, G. S., Zhou, X. H., Lin, Y., Yu, Q., Liu, S. R., Sun, W., Niu, S. L., Li, S. G., Zhang, J. H., and Yu, G. R.: Ecosystem traits linking functional traits to macroecology, Trends Ecol. Evol., 34, 200–210, https://doi.org/10.1016/j.tree.2018.11.004, 2019.
He, N. P., Liu, Y., Liu, C. C., Xu, L., Li, M. X., Zhang, J. H., He, J. S., Tang, Z. Y., Han, X. G., Ye, Q., Xiao, C. W., Yu, Q., Liu, S. R., Sun, W., Niu, S. L., Li, S. G., Sack, L., and Yu, G. R.: Plant trait networks: improved resolution of the dimensionality of adaptation, Trends Ecol. Evol., 35, 908–918, https://doi.org/10.1016/j.tree.2020.06.003, 2020.
Hutchinson, M. F. and Xu, T. B.: ANUSPLIN Version 4.4 User Guide, Fenner School of Environment and Society, the Australian National University, Canberra, 2013.
Integrated Scientific Expedition to Qinghai-Tibet Plateau, Chinese Academy of Sciences: Flora Xizangica, 5 volumes, Science Press, Beijing, 1983–1987.
Iversen, C. M., McCormack, M. L., Powell, A. S., Blackwood, C. B., Freschet, G. T., Kattge, J., Roumet, C., Stover, D. B., Soudzilovskaia, N. A., Valverde-Barrantes, O. J., van Bodegom, P. M., and Violle, C.: A global Fine-Root Ecology Database to address below-ground challenges in plant ecology, New Phytol., 215, 15–26, https://doi.org/10.1111/nph.14486, 2017.
Jin, Y., Wang, H., Xia, J., Ni, J., Li, K., Hou, Y., Hu, J., Wei, L., Xia, H., and Zhou, B.: A dataset of leaf traits on the Tibetan Plateau (2018–2021), National Tibetan Plateau Data Center [data set], https://doi.org/10.11888/Terre.tpdc.272516, 2022a.
Jin, Y. L., Wang, H. Y., Wei, L. F., Hu, J., Wu, K., Xia, H. J., Xia, J., Zhou, B. R., Li, K., and Ni, J.: A plot dataset of plant community of Qingzang Plateau, Chin. J. Plant Ecol., 46, 846–854, https://doi.org/10.17521/cjpe.2022.0174, 2022b.
Kattge, J., Díaz, S., Lavorel, S., Prentice, I. C., Leadley, P., Bönisch, G., Garnier, E., Westoby, M., Reich, P. B., Wright, I. J., Cornelissen, J. H. C., Violle, C., Harrison, S. P., van Bodegom, P. M., Reichstein, M., Enquist, B. J., Soudzilovskaia, N. A., Ackerly, D. D., Anand, M., Atkin, O., Bahn, M., Baker, T. R., Baldocchi, D., Bekker, R., Blanco, C. C., Blonder, B., Bond, W. J., Bradstock, R., Bunker, D. E., Casanoves, F., Cavender-Bares, J., Chambers, J. Q., Chapin, F. S., Chave, J., Coomes, D., Cornwell, W. K., Craine, J. M., Dobrin, B. H., Duarte, L., Durka, W., Elser, J., Esser, G., Estiarte, M., Fagan, W. F., Fang, J., Fernandez-Mendez, F., Fidelis, A., Finegan, B., Flores, O., Ford, H., Frank, D., Freschet, G. T., Fyllas, N. M., Gallagher, R V.,. Green, W. A., Gutierrez, A. G., Hickler, T., Higgins, S. I., Hodgson, J. G., Jalili, A., Jansen, S., Joly, C. A., Kerkhoff, A. J., Kirkup, D., Kitajima, K., Kleyer, M., Klotz, S., Knops, J. M. H., Kramer, K., Kuhn, I., Kurokawa, H., Laughlin, D., Lee, T. D., Leishman, M., Lens, F., Lenz, T., Lewis, S. L., Lloyd, J., Llusia, J., Louault, F., Ma, S., Mahecha, M. D., Manning, P., Massad, T., Medlyn, B. E., Messier, J., Moles, A. T., Muller, S. C., Nadrowski, K., Naeem, S., Niinemets, U., Nollert, S., Nuske, A., Ogaya, R., Oleksyn, J., Onipchenko, V. G., Onoda, Y., Ordonez, J., Overbeck, G., Ozinga, W. A., Patino, S., Paula, S., Pausas, J. G., Penuelas, J., Phillips, O. L., Pillar, V., Poorter, H., Poorter, L., Poschlod, P., Prinzing, A., Proulx, R., Rammig, A., Reinsch, S., Reu, B., Sack, L., Salgado-Negre, B., Sardans, J., Shiodera, S., Shipley, B., Siefert, A., Sosinski, E., Soussana, J. F., Swaine, E., Swenson, N., Thompson, K., Thornton, P., Waldram, M., Weiher, E., White, M., White, S., Wright, S. J., Yguel, B., Zaehle, S., Zanne, A. E., and Wirth, C.: TRY – a global database of plant traits, Global Change Biol., 17, 2905–2935, https://doi.org/10.1111/j.1365-2486.2011.02451.x, 2011.
Kattge, J., Bonisch, G., Diaz, S., Lavorel, S., Prentice, I. C., Leadley, P., Tautenhahn, S., Werner, G. D. A., Aakala, T., Abedi, M., Acosta, A. T. R., Adamidis, G. C., Adamson, K., Aiba, M., Albert, C. H., Alcantara, J. M., Alcazar, C. C., Aleixo, I., Ali, H., Amiaud, B., Ammer, C., Amoroso, M. M., Anand, M., Anderson, C., Anten, N., Antos, J., Apgaua, D. M. G., Ashman, T. L., Asmara, D. H., Asner, G. P., Aspinwall, M., Atkin, O., Aubin, I., Baastrup-Spohr, L., Bahalkeh, K., Bahn, M., Baker, T., Baker, W. J., Bakker, J. P., Baldocchi, D., Baltzer, J., Banerjee, A., Baranger, A., Barlow, J., Barneche, D. R., Baruch, Z., Bastianelli, D., Battles, J., Bauerle, W., Bauters, M., Bazzato, E., Beckmann, M., Beeckman, H., Beierkuhnlein, C., Bekker, R., Belfry, G., Belluau, M., Beloiu, M., Benavides, R., Benomar, L., Berdugo-Lattke, M. L., Berenguer, E., Bergamin, R., Bergmann, J., Carlucci, M. B., Berner, L., Bernhardt-Romermann, M., Bigler, C., Bjorkman, A. D., Blackman, C., Blanco, C., Blonder, B., Blumenthal, D., Bocanegra-Gonzalez, K. T., Boeckx, P., Bohlman, S., Bohning-Gaese, K., Boisvert-Marsh, L., Bond, W., Bond-Lamberty, B., Boom, A., Boonman, C. C. F., Bordin, K., Boughton, E. H., Boukili, V., Bowman, D. M. J. S., Bravo, S., Brendel, M. R., Broadley, M. R., Brown, K. A., Bruelheide, H., Brumnich, F., Bruun, H. H., Bruy, D., Buchanan, S. W., Bucher, S. F., Buchmann, N., Buitenwerf, R., Bunker, D. E., Burger, J., Burrascano, S., Burslem, D. F. R. P., Butterfield, B. J., Byun, C., Marques, M., Scalon, M. C., Caccianiga, M., Cadotte, M., Cailleret, M., Camac, J., Camarero, J. J., Campany, C., Campetella, G., Campos, J. A., Cano-Arboleda, L., Canullo, R., Carbognani, M., Carvalho, F., Casanoves, F., Castagneyrol, B., Catford, J. A., Cavender-Bares, J., Cerabolini, B. E. L., Cervellini, M., Chacon-Madrigal, E., Chapin, K., Chapin, F. S., Chelli, S., Chen, S. C., Chen, A. P., Cherubini, P., Chianucci, F., Choat, B., Chung, K. S., Chytry, M., Ciccarelli, D., Coll, L., Collins, C. G., Conti, L., Coomes, D., Cornelissen, J. H. C., Cornwell, W. K., Corona, P., Coyea, M., Craine, J., Craven, D., Cromsigt, J. P. G. M., Csecserits, A., Cufar, K., Cuntz, M., da Silva, A. C., Dahlin, K. M., Dainese, M., Dalke, I., Dalle Fratte, M., Anh, T. D. L., Danihelka, J., Dannoura, M., Dawson, S., de Beer, A. J., De Frutos, A., De Long, J. R., Dechant, B., Delagrange, S., Delpierre, N., Derroire, G., Dias, A. S., Diaz-Toribio, M. H., Dimitrakopoulos, P. G., Dobrowolski, M., Doktor, D., Drevojan, P., Dong, N., Dransfield, J., Dressler, S., Duarte, L., Ducouret, E., Dullinger, S., Durka, W., Duursma, R., Dymova, O., E-Vojtko, A., Eckstein, R. L., Ejtehadi, H., Elser, J., Emilio, T., Engemann, K., Erfanian, M. B., Erfmeier, A., Esquivel-Muelbert, A., Esser, G., Estiarte, M., Domingues, T. F., Fagan, W. F., Fagundez, J., Falster, D. S., Fan, Y., Fang, J. Y., Farris, E., Fazlioglu, F., Feng, Y. H., Fernandez-Mendez, F., Ferrara, C., Ferreira, J., Fidelis, A., Finegan, B., Firn, J., Flowers, T. J., Flynn, D. F. B., Fontana, V., Forey, E., Forgiarini, C., Francois, L., Frangipani, M., Frank, D., Frenette-Dussault, C., Freschet, G. T., Fry, E. L., Fyllas, N. M., Mazzochini, G. G., Gachet, S., Gallagher, R., Ganade, G., Ganga, F., Garcia-Palacios, P., Gargaglione, V., Garnier, E., Garrido, J. L., de Gasper, A. L., Gea-Izquierdo, G., Gibson, D., Gillison, A. N., Giroldo, A., Glasenhardt, M. C., Gleason, S., Gliesch, M., Goldberg, E., Goldel, B., Gonzalez-Akre, E., Gonzalez-Andujar, J. L., Gonzalez-Melo, A., Gonzalez-Robles, A., Graae, B. J., Granda, E., Graves, S., Green, W. A., Gregor, T., Gross, N., Guerin, G. R., Gunther, A., Gutierrez, A. G., Haddock, L., Haines, A., Hall, J., Hambuckers, A., Han, W. X., Harrison, S. P., Hattingh, W., Hawes, J. E., He, T. H., He, P. C., Heberling, J. M., Helm, A., Hempel, S., Hentschel, J., Herault, B., Heres, A. M., Herz, K., Heuertz, M., Hickler, T., Hietz, P., Higuchi, P., Hipp, A. L., Hirons, A., Hock, M., Hogan, J. A., Holl, K., Honnay, O., Hornstein, D., Hou, E. Q., Hough-Snee, N., Hovstad, K. A., Ichie, T., Igic, B., Illa, E., Isaac, M., Ishihara, M., Ivanov, L., Ivanova, L., Iversen, C. M., Izquierdo, J., Jackson, R. B., Jackson, B., Jactel, H., Jagodzinski, A. M., Jandt, U., Jansen, S., Jenkins, T., Jentsch, A., Jespersen, J. R. P., Jiang, G. F., Johansen, J. L., Johnson, D., Jokela, E. J., Joly, C. A., Jordan, G. J., Joseph, G. S., Junaedi, D., Junker, R. R., Justes, E., Kabzems, R., Kane, J., Kaplan, Z., Kattenborn, T., Kavelenova, L., Kearsley, E., Kempel, A., Kenzo, T., Kerkhoff, A., Khalil, M. I., Kinlock, N. L., Kissling, W. D., Kitajima, K., Kitzberger, T., Kjoller, R., Klein, T., Kleyer, M., Klimesova, J., Klipel, J., Kloeppel, B., Klotz, S., Knops, J. M. H., Kohyama, T., Koike, F., Kollmann, J., Komac, B., Komatsu, K., Konig, C., Kraft, N. J. B., Kramer, K., Kreft, H., Kuhn, I., Kumarathunge, D., Kuppler, J., Kurokawa, H., Kurosawa, Y., Kuyah, S., Laclau, J. P., Lafleur, B., Lallai, E., Lamb, E., Lamprecht, A., Larkin, D. J., Laughlin, D., Le Bagousse-Pinguet, Y., le Maire, G., le Roux, P. C., le Roux, E., Lee, T., Lens, F., Lewis, S. L., Lhotsky, B., Li, Y. Z., Li, X. E., Lichstein, J. W., Liebergesell, M., Lim, J. Y., Lin, Y. S., Linares, J. C., Liu, C. J., Liu, D. J., Liu, U., Livingstone, S., Llusia, J., Lohbeck, M., Lopez-Garcia, A., Lopez-Gonzalez, G., Lososova, Z., Louault, F., Lukacs, B. A., Lukes, P., Luo, Y. J., Lussu, M., Ma, S. Y., Pereira, C. M. R., Mack, M., Maire, V., Makela, A., Makinen, H., Malhado, A. C. M., Mallik, A., Manning, P., Manzoni, S., Marchetti, Z., Marchino, L., Marcilio-Silva, V., Marcon, E., Marignani, M., Markesteijn, L., Martin, A., Martinez-Garza, C., Martinez-Vilalta, J., Maskova, T., Mason, K., Mason, N., Massad, T. J., Masse, J., Mayrose, I., McCarthy, J., McCormack, M. L., McCulloh, K., McFadden, I. R., McGill, B. J., McPartland, M. Y., Medeiros, J. S., Medlyn, B., Meerts, P., Mehrabi, Z., Meir, P., Melo, F. P. L., Mencuccini, M., Meredieu, C., Messier, J., Meszaros, I., Metsaranta, J., Michaletz, S. T., Michelaki, C., Migalina, S., Milla, R., Miller, J. E. D., Minden, V., Ming, R., Mokany, K., Moles, A. T., Molnar, V. A., Molofsky, J., Molz, M., Montgomery, R. A., Monty, A., Moravcova, L., Moreno-Martinez, A., Moretti, M., Mori, A. S., Mori, S., Morris, D., Morrison, J., Mucina, L., Mueller, S., Muir, C. D., Muller, S. C., Munoz, F., Myers-Smith, I. H., Myster, R. W., Nagano, M., Naidu, S., Narayanan, A., Natesan, B., Negoita, L., Nelson, A. S.,. Neuschulz, E. L., Ni, J., Niedrist, G., Nieto, J., Niinemets, U, Nolan, R., Nottebrock, H., Nouvellon, Y., Novakovskiy, A., Nystuen, K. O., O'Grady, A., O'Hara, K., O'Reilly-Nugent, A., Oakley, S., Oberhuber, W., Ohtsuka, T., Oliveira, R., Ollerer, K., Olson, M. E., Onipchenko, V., Onoda, Y., Onstein, R. E., Ordonez, J. C., Osada, N., Ostonen, I., Ottaviani, G., Otto, S., Overbeck, G. E., Ozinga, W. A., Pahl, A. T., Paine, C. E. T., Pakeman, R. J., Papageorgiou, A. C., Parfionova, E., Partel, M., Patacca, M., Paula, S., Paule, J., Pauli, H., Pausas, J. G., Peco, B., Penuelas, J., Perea, A., Peri, P. L., Petisco-Souza, A. C., Petraglia, A., Petritan, A. M., Phillips, O. L., Pierce, S., Pillar, V. D., Pisek, J., Pomogaybin, A., Poorter, H., Portsmuth, A., Poschlod, P., Potvin, C., Pounds, D., Powell, A. S., Power, S. A., Prinzing, A., Puglielli, G., Pysek, P., Raevel, V., Rammig, A., Ransijn, J., Ray, C. A., Reich, P. B., Reichstein, M., Reid, D. E. B., Rejou-Mechain, M., de Dios, V. R., Ribeiro, S., Richardson, S., Riibak, K., Rillig, M. C., Riviera, F., Robert, E. M. R., Roberts, S., Robroek, B., Roddy, A., Rodrigues, A. V., Rogers, A., Rollinson, E., Rolo, V., Romermann, C., Ronzhina, D., Roscher, C., Rosell, J. A., Rosenfield, M. F., Rossi, C., Roy, D. B., Royer-Tardif, S., Ruger, N., Ruiz-Peinado, R., Rumpf, S. B., Rusch, G. M., Ryo, M., Sack, L., Saldana, A., Salgado-Negret, B., Salguero-Gomez, R., Santa-Regina, I., Santacruz-Garcia, A. C., Santos, J., Sardans, J., Schamp, B., Scherer-Lorenzen, M., Schleuning, M., Schmid, B., Schmidt, M., Schmitt, S., Schneider, J. V., Schowanek, S. D., Schrader, J., Schrodt, F., Schuldt, B., Schurr, F., Garvizu, G. S., Semchenko, M., Seymour, C., Sfair, J. C., Sharpe, J. M., Sheppard, C. S., Sheremetiev, S., Shiodera, S., Shipley, B., Shovon, T. A., Siebenkas, A., Sierra, C., Silva, V., Silva, M., Sitzia, T., Sjoman, H., Slot, M., Smith, N. G., Sodhi, D., Soltis, P., Soltis, D., Somers, B., Sonnier, G., Sorensen, M. V., Sosinski, E. E., Soudzilovskaia, N. A., Souza, A. F., Spasojevic, M., Sperandii, M. G., Stan, A. B., Stegen, J., Steinbauer, K., Stephan, J. G., Sterck, F., Stojanovic, D. B., Strydom, T., Suarez, M. L., Svenning, J. C., Svitkova, I., Svitok, M., Svoboda, M., Swaine, E., Swenson, N., Tabarelli, M., Takagi, K., Tappeiner, U., Tarifa, R., Tauugourdeau, S., Tavsanoglu, C., te Beest, M., Tedersoo, L., Thiffault, N., Thom, D., Thomas, E., Thompson, K., Thornton, P. E., Thuiller, W., Tichy, L., Tissue, D., Tjoelker, M. G., Tng, D. Y. P., Tobias, J., Torok, P., Tarin, T., Torres-Ruiz, J. M., Tothmeresz, B., Treurnicht, M., Trivellone, V., Trolliet, F., Trotsiuk, V., Tsakalos, J. L., Tsiripidis, I., Tysklind, N., Umehara, T., Usoltsev, V., Vadeboncoeur, M., Vaezi, J., Valladares, F., Vamosi, J., van Bodegom, P. M., van Breugel, M., Van Cleemput, E., van de Weg, M., van der Merwe, S., van der Plas, F., van der Sande, M. T., van Kleunen, M., Van Meerbeek, K., Vanderwel, M., Vanselow, K. A., Varhammar, A., Varone, L., Valderrama, M. Y., Vassilev, K., Vellend, M., Veneklaas, E. J., Verbeeck, H., Verheyen, K., Vibrans, A., Vieira, I., Villacis, J., Violle, C., Vivek, P., Wagner, K., Waldram, M., Waldron, A., Walker, A. P., Waller, M., Walther, G., Wang, H., Wang, F., Wang, W. Q., Watkins, H., Watkins, J., Weber, U., Weedon, J. T., Wei, L. P., Weigelt, P., Weiher, E., Wells, A. W., Wellstein, C., Wenk, E., Westoby, M., Westwood, A., White, P. J., Whitten, M., Williams, M., Winkler, D. E., Winter, K., Womack, C., Wright, I. J., Wright, S. J., Wright, J., Pinho, B. X., Ximenes, F., Yamada, T., Yamaji, K., Yanai, R., Yankov, N., Yguel, B., Zanini, K. J., Zanne, A. E., Zeleny, D., Zhao, Y. P., Zheng, J. M., Zheng, J., Zieminska, K., Zirbel, C. R., Zizka, G., Zo-Bi, I. C., Zotz, G., and Wirth, C.: TRY plant trait database – enhanced coverage and open access, Global Change Biol., 26, 119–188, https://doi.org/10.1111/gcb.14904, 2020.
Liu, Y. X.: Flora in Desertis Reipublicae Populorum Sinarum, 3 volumes, Science Press, Beijing, 1985–1992.
Lu, L. M., Mao, L. F., Yang, T., Ye, J. F., Liu, B., Li, H. L., Sun, M., Miller, J. T., Mathews, S., Hu, H. H., Niu, Y. T., Peng, D. X., Chen, Y. H., Smith, S. A., Chen, M., Xiang, K. L., Le, C. T., Dang, V. C., Lu, A. M., Soltis, P. S., Soltis, D. E., Li, J. H., and Chen, Z. D.: Evolutionary history of the angiosperm flora of China, Nature, 554, 234–238, https://doi.org/10.1038/nature25485, 2018.
Luo, T. X., Luo, J., and Pan, Y. D.: Leaf traits and associated ecosystem characteristics across subtropical and timberline forests in the Gongga Mountains, Eastern Tibetan Plateau, Oecologia, 142, 261–273, https://doi.org/10.1007/s00442-004-1729-6, 2005.
Ma, Z. Q., Guo, D. L., Xu, X. L., Lu, M. Z., Bardgett, R. D., Eissenstat, D. M., McCormack, M. L., and Hedin, L. O.: Evolutionary history resolves global organization of root functional traits, Nature, 555, 94–97, https://doi.org/10.1038/nature26163, 2018.
Maes, S. L., Perring, M. P., Depauw, L., Bernhardt-Romermann, M., Blondeel, H., Brumelis, G., Brunet, J., Decocq, G., den Ouden, J., Govaert, S., Hardtle, W., Hedl, R., Heinken, T., Heinrichs, S., Hertzog, L., Jaroszewicz, B., Kirby, K., Kopecky, M., Landuyt, D., Malis, F., Vanneste, T., Wulf, M., and Verheyen, K.: Plant functional trait response to environmental drivers across European temperate forest understorey communities, Plant Biol., 22, 410–424, https://doi.org/10.1111/plb.13082, 2020.
Mariano, E., Gomes, T. F., Lins, S. R. M., Abdalla- Filho, A. L., Soltangheisi, A., Araújo, M. G. S., Almeida, R. F., Augusto, F. G., Canisares, L. P., Chaves, S. S. F., Costa, C. F. G., Diniz-Reis, T. R., Galera, L. A., Martinez, M. G., Morais, M. C., Perez, E. B., Reis, L. C., Simon, C. D., Mardegan, S. F., Domingues, T. F., Miatto, R. C., Oliveira, R. S., Reis, C. R. G., Nardoto, G. B., Kattge, J., and Martinelli, L. A.: LT-Brazil: A database of leaf traits across biomes and vegetation types in Brazil, Global Ecol. Biogeogr., 30, 2136–2146, https://doi.org/10.1111/geb.13381, 2021.
Meng, T. T., Ni, J., and Harrison, S. P.: Plant morphometric traits and climate gradients in northern China: a meta-analysis using quadrat and flora data, Ann. Bot., 104, 1217–1229, https://doi.org/10.1093/aob/mcp230, 2009.
Meng, T.-T., Wang, H., Harrison, S. P., Prentice, I. C., Ni, J., and Wang, G.: Responses of leaf traits to climatic gradients: adaptive variation versus compositional shifts, Biogeosciences, 12, 5339–5352, https://doi.org/10.5194/bg-12-5339-2015, 2015.
Moles, A. T., Ackerly, D. D., Tweddle, J. C., Dickie, J. B., Smith, R., Leishman, M. R., Mayfield, M. M., Pitman, A., Wood, J. T., and Westoby, M.: Global patterns in seed size, Global Ecol. Biogeogr., 16, 109–116, https://doi.org/10.1111/j.1466-8238.2006.00259.x, 2007.
Moles, A. T., Warton, D. I., Warman, L., Swenson, N. G., Laffan, S. W., Zanne, A. E., Pitman, A., Hemmings, F. A., and Leishman, M. R.: Global patterns in plant height, J. Ecol., 97, 923–932, https://doi.org/10.1111/j.1365-2745.2009.01526.x, 2009.
Myers-Smith, I. H. Thomas, H. J. D., and Bjorkman, A. D.: Plant traits inform predictions of tundra responses to global change, New Phytol., 221, 1742–1748, https://doi.org/10.1111/nph.15592, 2019.
Pérez-Harguindeguy, N., Díaz, S., Garnier, E., Lavorel, S., Poorter, H., Jaureguiberry, P., Bret-Harte, M. S., Cornwell, W. K., Craine, J. M., Gurvich, D. E., Urcelay, C., Veneklaas, E. J., Reich, P. B., Poorter, L., Wright, I. J., Ray, P., Enrico, L., Pausas, J. G., de Vos, A. C., Buchmann, N., Funes, G., Quétier, F., Hodgson, J. G., Thompson, K., Morgan, H. D., ter Steege, H., van der Heijden, M. G. A., Sack, L., Blonder, B., Poschlod, P., Vaieretti, M. V., Conti, G., Staver, A. C., Aquino, S., and Cornelissen, J. H. C.: New handbook for standardised measurement of plant functional traits worldwide, Aust. J. Bot., 61, 167–234, https://doi.org/10.1071/BT12225, 2013.
Piao, S. L., Zhang, X. Z., Wang, T., Liang, E. Y., Wang, S. P., Zhu, J. T., and Niu, B.: Responses and feedback of the Tibetan Plateau's alpine ecosystem to climate change, Chin. Sci. Bull., 64, 2842–2855, https://doi.org/10.1360/TB-2019-0074, 2019.
Reich, P. B. and Oleksyn, J.: Global patterns of plant leaf N and P in relation to temperature and latitude, P. Natl. Acad. Sci. USA, 101, 11001–11006, https://doi.org/10.1073/pnas.0403588101, 2004.
Reichstein, M., Bahn, M., Mahecha, M. D., Kattge, J., and Baldocchi, D. D.: Linking plant and ecosystem functional biogeography, P. Natl. Acad. Sci. USA, 111, 13697–13702, https://doi.org/10.1073/pnas.1216065111, 2014.
Roddy, A. B., Martínez-Perez, C., Teixido, A. L., Cornelissen, T. G., Olson, M. E., Oliveira, R. S., and Silveira, F. A. O.: Towards the flower economics spectrum, New Phytol., 229, 665–672, https://doi.org/10.1111/nph.16823, 2021.
Shi, W. Q., Wang, G. A., and Han, W. X.: Altitudinal variation in leaf nitrogen concentration on the eastern slope of Mount Gongga on the Tibetan Plateau, China, Plos One, 7, e44628, https://doi.org/10.1371/journal.pone.0044628, 2012.
Sloan, S., Jenkins, C. N., Joppa, L. N., Gaveau, D. L. A., and Laurance, W. F.: Remaining natural vegetation in the global biodiversity hotspots, Biol. Conserv., 177, 12–24, https://doi.org/10.1016/j.biocon.2014.05.027, 2014.
Taseski, G. M., Beloe, C. J., Gallagher, R. V., Chan, J. Y., Dalrymple, R. L., and Cornwell, W. K.: A global-growth form database for 143,616 vascular plant species, Ecology, 100, e02614, https://doi.org/10.1002/ecy.2614, 2019.
Tavşanoğlu, Ç., and Pausas, J. G.: A functional trait database for Mediterranean Basin plants, Sci. Data, 5, 180135, https://doi.org/10.1038/sdata.2018.135, 2018.
van Bodegom, P. M., Douma, J. C., and Verheijen, L. M.: A fully traits-based approach to modeling global vegetation distribution, P. Natl. Acad. Sci. USA, 111, 13733–13738, https://doi.org/10.1073/pnas.1304551110, 2014.
Vandvik, V., Halbritter, A. H., Yang, Y., He, H., Zhang, L., Brummer, A. B., Klanderud, K., Maitner, B. S., Michaletz, S. T., Sun, X. Y., Telford, R. J., Wang, G. X., Althuizen, I. H. J., Henn, J. J., Garcia, W. F. E., Gya, R., Jaroszynska, F., Joyce, B. L., Lehman, R., Moerland, M. S., Nesheim-Hauge, E., Nordås, L. H., Peng, A., Ponsac, C., Seltzer, L., Steyn, C., Sullivan, M. K., Tjendra, J., Xiao, Y., Zhao, X. X., and Enquist, B. J.: Plant traits and vegetation data from climate warming experiments along an 1100 m elevation gradient in Gongga Mountains, China, Sci. Data, 7, 189, https://doi.org/10.1038/s41597-020-0529-0, 2020.
Violle, C., Navas, M. L., Vile, D., Kazakou, E., Fortunel, C., Hummel, I., and Garnier, E.: Let the concept of trait be functional!, Oikos, 116, 882–892, https://doi.org/10.1111/j.0030-1299.2007.15559.x, 2007.
Wang, C. S., Lyu, W. W., Jiang, L. L., Wang, S. P., Wang, Q., Meng, F. D., and Zhang, L. R.: Changes in leaf vein traits among vein types of alpine grassland plants on the Tibetan Plateau, J. Mt. Sci., 17, 2161–2169, https://doi.org/10.1007/s11629-020-6069-4, 2020.
Wang, H., Prentice, I. C., Keenan, T. F., Davis, T. W., Wright, I. J., Cornwell, W. K., Evans, B. J., and Peng, C. H.: Towards a universal model for carbon dioxide uptake by plants, Nat. Plants, 3, 734–741, https://doi.org/10.1038/s41477-017-0006-8, 2017.
Wang, H., Harrison, S. P., Prentice, I. C., Yang, Y. Z., Bai, F., Togashi, H. F., Wang, M., Zhou, S. X., and Ni, J.: The China Plant Trait Database: toward a comprehensive regional compilation of functional traits for land plants, Ecology, 99, p. 500, https://doi.org/10.1002/ecy.2091, 2018.
Wang, H., Wang, R. X., Harrison, S. P., and Prentice, I. C.: Leaf morphological traits as adaptations to multiple climate gradients, J. Ecol., 110, 1344–1355, https://doi.org/10.1111/1365-2745.13873, 2022.
Wang, Q. and Hong, D. Y.: Understanding the plant diversity on the roof of the world, The Innovation, 3, 100215, https://doi.org/10.1016/j.xinn.2022.100215, 2022.
Wei, L. F., Hu, X. F., Cheng, Q., Wu, X. Q., and Ni, J.: A dataset of spatial distribution of bioclimatic variables inChina at 1 km resolution, China Sci. Data [data set], https://doi.org/10.11922/11-6035.csd.2022.0003.zh, 2022.
Weigelt, P., König, C., and Kreft, H.: GIFT – A Global Inventory of Floras and Traits for macroecology and biogeography, J. Biogeogr., 47, 16–43, https://doi.org/10.1111/jbi.13623, 2019.
Wright, I. J., Reich, P. B., Westoby, M., Ackerly, D. D., Baruch, Z., Bongers, F., Cavender-Bares, J., Chapin, T., Cornelissen, J. H. C., Diemer, M., Flexas, J., Garnier, E., Groom, P. K., Gulias, J., Hikosaka, K., Lamont, B. B., Lee, T., Lee, W., Lusk, C., Midgley, J. J., Navas, M. L., Niinemets, Ü., Oleksyn, J., Osada, N., Poorter, H., Poot, P., Prior, L., Pyankov, V. I., Roumet, C., Thomas, S. C., Tjoelker, M. G., Veneklaas, E. J., and Villar, R.: The worldwide leaf economics spectrum, Nature, 428, 821–827, https://doi.org/10.1038/nature02403, 2004.
Wu, D. X.: Protocols for Standard Biological Observation and Measurement in Terrestrial Ecosystems, China Environmental Science Press, Beijing, ISBN 9787511141071, 2007.
Wullschleger, S. D., Epstein, H. E., Box, E. O., Euskirchen, E. S., Goswami, S., Iversen, C. M., Kattge, J., Norby, R. J., van Bodegom, P. M., and Xu, X. F.: Plant functional types in Earth system models: past experiences and future directions for application of dynamic vegetation models in high-latitude ecosystems, Ann. Bot., 114, 1–16, https://doi.org/10.1093/aob/mcu077, 2014.
Xu, H. Y., Wang, H., Prentice, I. C., Harrison, S. P., Wang, G. X., and Sun, X. Y.: Predictability of leaf traits with climate and elevation: a case study in Gongga Mountain, China, Tree Physiol., 41, 13360–1352, https://doi.org/10.1093/treephys/tpab003, 2021.
Xu, T. B. and Hutchinson, M. F.: New developments and applications in the ANUCLIM spatial climatic and bioclimatic modelling package, Environ. Modell. Softw., 40, 267–279, https://doi.org/10.1016/j.envsoft.2012.10.003, 2013.
Yan, Y. J., Yang, X., and Tang, Z. Y.: Patterns of species diversity and phylogenetic structure of vascular plants on the Qinghai-Tibetan Plateau, Ecol. Evol., 3, 4584–4595, https://doi.org/10.1002/ece3.847, 2013.
Yao, T. D., Thompson, L., Yang, W., Yu, W. S., Gao, Y., Guo, X. J., Yang, X. X., Duan, K. Q., Zhao, H. B., Xu, B. Q., Pu, J. C., Lu, A. X., Xiang, Y., Kattel, D. B., and Joswiak, D.: Different glacier status with atmospheric circulations in Tibetan Plateau and surroundings, Nat. Clim. Change, 2, 663–667, https://doi.org/10.1038/nclimate1580, 2012.
Zhang, X. Z., Yang, Y. P., Piao, S. L., Bao, W. K., Wang, S. P., Wang, G. X., Sun, H., Luo, T. X., Zhang, Y. J., Shi, P. L., Liang, E. Y., Shen, M. G., Wang, J. S., Gao, Q. Z., Zhang, Y. L., and Ouyang, H.: Ecological change on the Tibetan Plateau, Chin. Sci. Bull., 60, 3048–3056, https://doi.org/10.1360/N972014-01339, 2015.
- Abstract
- Introduction
- Study areas
- Materials and methods
- Data description of sampling sites
- Data description of species and traits
- Data availability
- Summary
- Author contributions
- Competing interests
- Disclaimer
- Special issue statement
- Acknowledgements
- Financial support
- Review statement
- References
- Supplement
- Abstract
- Introduction
- Study areas
- Materials and methods
- Data description of sampling sites
- Data description of species and traits
- Data availability
- Summary
- Author contributions
- Competing interests
- Disclaimer
- Special issue statement
- Acknowledgements
- Financial support
- Review statement
- References
- Supplement