the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Best practices for data management in marine science: lessons from the Nansen Legacy project
Øystein Godøy
Tove Margrethe Gabrielsen
Pål Gunnar Ellingsen
Marit Reigstad
Miriam Marquardt
Arnfinn Morvik
Helge Sagen
Stein Tronstad
Lara Ferrighi
Large, multidisciplinary projects that collect vast amounts of data are becoming increasingly common in academia. Efficiently managing data across and beyond such projects necessitates a shift from fragmented efforts to coordinated, collaborative approaches. This article presents the data management strategies employed in the Nansen Legacy project (https://doi.org/10.1016/B978-0-323-90427-8.00009-5, Wassmann, 2022), a multidisciplinary Norwegian research initiative involving over 300 researchers and 20 expeditions into and around the northern Barents Sea. To enhance consistency in data collection, sampling protocols were developed and implemented across different teams and expeditions. A searchable metadata catalogue was established, providing an overview of all collected data within weeks of each expedition. The project also implemented a policy that mandates immediate data sharing among members and publishing of data in accordance with the FAIR guiding principles where feasible. We detail how these strategies were implemented and discuss the successes and challenges, offering insights and lessons learned to guide future projects in similar endeavours.
- Article
(2396 KB) - Full-text XML
- BibTeX
- EndNote
We are now firmly in the age of big data, where datasets are so vast that they exceed the capacity of a single person or project to collect and process. These large datasets are crucial for addressing some of today's most pressing scientific questions. Data from diverse sources can be integrated into monitoring systems that track changes in our dynamic environment. Data provide the foundation for models that provide forecasts and projections of future changes and their impacts, serving as powerful tools for decision making.
To maximise the effectiveness of scientific research, a shift from fragmented efforts to coordinated, collaborative endeavours is essential. Central to this shift is the way data are managed and governed. This coordination should extend throughout the entire data workflow, ensuring that data collection methods are consistent, thus allowing datasets to be compared, synthesised and reused effectively, both within and beyond the project. Transparent tracking of data collection activities enables better coordination within and between projects, fostering the collection of complementary data and filling gaps rather than duplicating efforts. Early sharing and publication of data accelerate scientific progress, as data can be reused more rapidly. The FAIR guiding principles (Wilkinson et al., 2016) offer a framework on how to make data machine-actionable, enabling integration into services that benefit society.
The Nansen Legacy project (Wassmann, 2022) is a Norwegian research initiative aimed at understanding the profound changes observed in the Northern Barents Sea and the Arctic as a whole. Spanning from 2018 to 2024, this multidisciplinary project has conducted 20 extensive research expeditions, integrating a wide array of scientific disciplines, including oceanography, meteorology, marine biology, marine chemistry, geology and engineering. In this article, we will explore the challenges and opportunities associated with data management in such a large-scale, multidisciplinary project.
Effective data management was prioritised from the very start of the project, beginning with the preparation of the proposal. A dedicated team consisting data managers and scientists from all partner institutions, outlined the principles that would govern the project and incorporated these into the first draft of the project's data policy (The Nansen Legacy, 2021) and data management plan (The Nansen Legacy, 2024) at the outset. The leadership team's involvement was crucial in ensuring these foundational documents were both well-conceived and effectively implemented. These documents served as the cornerstone for all subsequent data management activities discussed in this paper.
The project allocated resources and competence through a dedicated work package on data management led with complementary expertise. This included experience from international data management structures (from e.g. World Meteorological Organization), genetic database systems and physical and biological field work. A dedicated full time data manager was appointed to plan, develop and support the data handling. In addition, a data management resource group with data managers from each partner institution was established to strengthen collaboration, facilitate harmonised handling across disciplines and institutions, and support a legacy beyond the project period. This may facilitate further development and cultural change. Project management provided funding for training and data publishing workshops to ensure broad involvement.
This article provides a comprehensive overview of the data management practices implemented throughout the Nansen Legacy project, ordered according to the typical data cycle (Fig. 1):
-
Consistent data collection. We begin by exploring the importance of standardising the data collection processes. This section details the implementation of sampling protocols designed to ensure consistency across various data collectors.
-
Keeping track of data collected. We examine how we monitored and documented the collection process. This includes methods for keeping both project members and external stakeholders informed about the data collected, including its location and timing.
-
Data storage and sharing within the project. The article addresses the storage of unpublished data, focusing on how storage solutions were developed to support efficient data sharing within the project while adhering to best practices in data security.
-
Data publishing. We discuss the publication process of the project's data, emphasising our efforts to adhere to the FAIR (Findable, Accessible, Interoperable, and Reusable) data management principles (Wilkinson et al., 2016) wherever feasible.
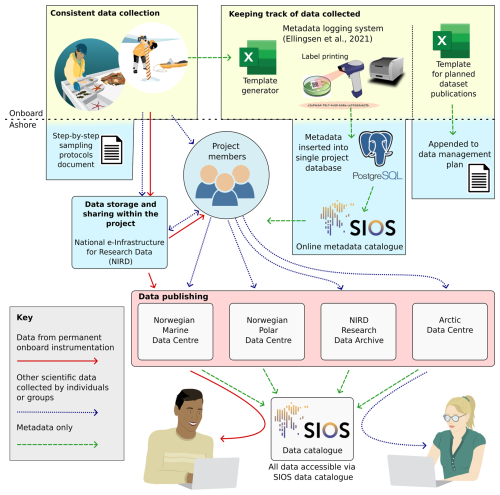
Figure 1The Nansen Legacy data workflow, encompassing all stages from data collection through to publication. The figure includes references to specific sections of the article (bold font) where each aspect of the data workflow is discussed in detail.
Each section is further divided into subsections that cover: (1) the motivations and objectives of the project related to each aspect of the data workflow, (2) the methods used to achieve these objectives, including the support provided by the data management team to facilitate these processes, and (3) an evaluation of outcomes, key lessons learned, and recommendations for the broader data management community and for future large-scale projects.
To clarify how the FAIR principles were considered throughout the project, each relevant aspect is annotated inline using the corresponding initial – F (Findable), A (Accessible), I (Interoperable), and R (Reusable) – shown in parentheses.
At the end of the article is a discussion and summary section that highlights the importance of cultural and organisational changes required for implementing and sustaining the data management practices within and beyond the Nansen Legacy project. This section also summarises the key findings outlined in the article.
2.1 Motivation and Aims
The Nansen Legacy project conducted 20 research expeditions across all seasons to produce multi-year time series. An important but sometimes overlooked aspect of data management is maintaining consistent data collection methods to ensure comparability across datasets.
2.2 Methods
The project collaboratively developed a series of sampling protocols – detailed, step-by-step instructions for collecting each type of data. The first step was to identify researchers interested in collecting similar types of samples. Researchers from different institutions agreed on the methodology for the specific sampling and analysis planned to ensure comparable data across different cruises and institutional responsibilities. Detailed protocols were developed collaboratively and published, with coordination led by a senior engineer who worked closely with each researcher and research group. They ensured that all methods and sampling strategies were properly described and that all relevant scientists were included in the preparations ahead of the cruises. Updates were made when new methods were included or improvements made, ultimately resulting in ten versions being published (e.g. The Nansen Legacy, 2022). By referring to these protocols in published datasets and data papers, the project enhances the transparency of its data collection processes, providing users with a clear understanding of how the data were gathered and processed (R). These protocols can be used to more effectively onboard new people into the project, and can be referred to when writing the methodology section of scientific articles (e.g. Marquardt et al., 2023b; Koenig et al., 2024).
2.3 Outcomes and Lessons Learned
The sampling protocols were widely adopted across the Nansen Legacy project and have even been utilised beyond the project, for example at the UiT the Arctic University of Norway, the University Centre in Svalbard and the University of Agder. The bottom-up approach, involving researchers directly in the design and implementation of these protocols, proved to be highly effective. We encourage future researchers and projects to use and build upon these protocols. Adopting shared methodologies is crucial for ensuring data can be reliably integrated across providers and projects. Without this, integration becomes difficult, potentially leading to measurement bias, gaps in understanding and undermining long-term monitoring efforts.
3.1 Motivation and Aims
In large-scale, multidisciplinary projects, effectively tracking data collection – what data was collected, by whom, where, and when – is crucial. Providing such an oversight not only facilitates cross-disciplinary collaboration but also enhances the reusability of the data by ensuring that it is well-documented and accessible for future use. This oversight further enables scientists to strategically plan future expeditions, focusing on complementary datasets and addressing potential gaps in coverage. Paired with the adoption of consistent sampling protocols, it can also reduce the environmental footprint of science by avoiding unnecessary duplication of observations within and between projects in the same timeline.
3.2 Methods
Within the Nansen Legacy project, addressing these challenges involved developing a metadata catalogue (Ellingsen et al., 2021) to provide an overview of all the data collected and maintaining an up-to-date data management plan (The Nansen Legacy, 2024). This metadata catalogue focuses on pre-publication metadata, capturing information that is part of the data production process rather than the final documentation and publishing of datasets. The data management plan includes an overview of all datasets that project participants plan to publish. The subsections below outline how these approaches were implemented.
3.2.1 Metadata Logging System
The metadata catalogue was developed to provide a searchable overview of all data collected during the expeditions (F, A). This catalogue includes only metadata descriptions and not the data themselves. The system is described in full in Ellingsen et al. (2021) and summarised below. Figure 2 presents an example workflow of a scientist using the metadata logging system.
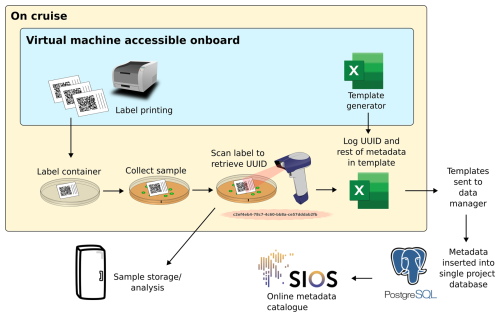
Figure 2An example workflow for the metadata logging system. The steps are not numbered as the order can vary between use cases. Steps include label printing, logging metadata in templates and combining all the templates into a searchable online metadata catalogue hosted at https://sios-svalbard.org/aen/tools, last access: 6 November 2025. Figure adapted from Ellingsen et al. (2021).
Ellingsen et al. (2021) developed a spreadsheet template generator to ensure consistent and structured metadata recording by all scientists. This template included required and recommended terms, ensuring all records contained essential information such as the collection date and location, the data collector, the principal investigator's contact details, and the type of sample (e.g., seawater sample, ice core, fish, virtual sample). Terms were taken from the Darwin Core terms (Darwin Core Community, 2010) or Climate and Forecast standard names (Eaton et al., 2022) where possible to encourage a consistent use of standard terms from data collection right through to publication (I, R). Other terms, not available in controlled vocabularies, were defined by the project to meet its specific needs.
Scientists could also reference the relevant section and version of the sampling protocols (discussed in Sect. 2) for detailed data collection procedures (R). Each metadata record was assigned a universally unique identifier (UUID), facilitating precise tracking (F). Label printers onboard the vessel produced labels with UUIDs encoded as scannable data matrices, linking physical samples to the electronic log. UUIDs can be generated using most common programming languages or using websites such as https://www.uuidgenerator.net, last access: 6 November 2025.
The metadata catalogue is hierarchical. The metadata for a sample can include the UUID of a “parent” record. For instance, if multiple fish were caught in a net, each fish would be recorded with its own UUID along with a reference to the net's parent UUID (see Fig. 3 for examples).
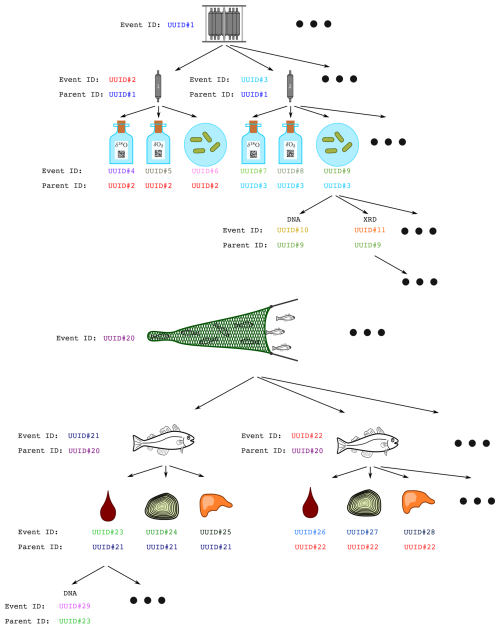
Figure 3The figure shows two examples of parent-child relation trees. Both trees display the inheritance of UUIDs from parent to child. Figure taken directly from Ellingsen et al. (2021).
Before the end of each cruise, the populated templates were verified using an onboard checker. Logs from all cruises were then combined into a PostgreSQL database – a free, open-source relational database management system. Shared metadata, such as time and coordinates, were propagated from parent to child records to ensure consistency. After each major update, the PostgreSQL table was exported as a new CSV file and made available as a searchable catalogue at https://sios-svalbard.org/aen/tools, last access: 6 November 2025 (F, A).
3.2.2 Planned Data Publications
On each cruise, the scientists listed each dataset they planned to publish using the data collected as an individual row in a shared spreadsheet template. Scientists included contact details for the principal investigator, estimated timeline for publication and details of any relevant embargo period requested for each dataset (The Nansen Legacy, 2021, policy VI). These tables were included in the project's data management plan (The Nansen Legacy, 2024), which has been revised through time. This was a useful resource for the project leadership and data management teams in tracking progress on data publication. The tables from each cruise have since been harmonised into a single table to provide an overview for the whole project (The Nansen Legacy, 2024), and data not collected on cruises (e.g. data output from models, long-term moorings or experiments) have been added.
3.3 Training and support
To ensure that metadata and planned data publication templates were filled in correctly, training webinars were held prior to each cruise or onboard. Members of the data management team participated in some research expeditions to offer support to scientists and to ensure there were no technical mishaps. They were contactable remotely on expeditions where they were not personally present. The data management team liaised with scientists following the cruises to fix any errors or for clarification on certain matters.
3.4 Outcomes and Lessons Learned
The uptake in filling out the templates thoroughly and accurately was very good, though it is difficult to quantify this precisely since unrecorded metadata remain unknown. For scientists with a large number of samples to log, this was a time-consuming process. Success was only possible thanks to the project leadership team fully committing to the process. Yet the procedure has been adopted by project alumni outside of the project, for example in student courses at the University Centre in Svalbard. In Nansen Legacy, the metadata catalogue includes 90 376 records from 490 spreadsheets. While many samples were labelled and logged correctly, in some cases a single label was assigned to a single bagged collection of samples. Some samples were recorded in the electronic log using UUIDs that did not correlate with the physical samples.
Future projects should aim to build logging systems that are less time-consuming. Using spreadsheet templates can be advantageous, as scientists are already familiar with them and do not need to learn a new system. However, projects with more time and resources for development could consider using dedicated software with a graphical user interface to simplify the logging process and automate tasks, especially when logging many samples with common metadata.
The link between physical samples and the electronic record in the metadata catalogue was also not complete. Scanning a sample's label would yield its UUID, requiring a separate search for the UUID in the metadata catalogue to retrieve its metadata. This could be improved by encoding a unique URL that includes the UUID for each sample within the metadata catalogue into the data matrix, enabling direct access via most smartphones. However, careful planning would be needed to determine where the metadata catalogue would be hosted and a defined pattern for each sample's URL in advance.
The metadata catalogue was widely used by interested parties both within and outside the project. This was useful in tracking down data as discussed in Sect. 4.4.
Keeping track of data not connected to a single research cruise proved more difficult. A complete overview of the project's data should also include data from long-term moorings, experiments and model outputs. This would require a single identifier for the project that would act as the parent for each child cruise and other data source. These were added to the data management plan on a case-by-case basis, but there was no formal routine for tracking these datasets.
4.1 Motivation and Aims
In a survey conducted by Tenopir et al. (2020), 85 % of respondents indicated their willingness to share their data with others and to use data collected by others if it were easily accessible. Despite this, more than half of the respondents admitted to following only “high” or “mediocre” risk practices for storing their data, such as on personal computers, departmental servers, or USB drives. Such practices can impede data sharing and expose data to security risks or the risk of losing the data. These results illustrate that whilst data security and sharing are prevalent issues, there is an encouraging willingness to improve. However, we strongly agree with the statement by Tenopir et al. (2020) that guidance from data managers is clearly needed to achieve this change.
Recognising this, the Nansen Legacy board implemented a policy mandating that all data, regardless of any embargo period, be made available to all project participants (The Nansen Legacy, 2021, policy IX). However, permission from the relevant principal investigator was required before any use of the data. This policy aimed to foster collaboration between research groups by providing early access to data, thereby enabling research to progress with minimal delay.
4.2 Methods
Standard cruise data from onboard instrumentation was transferred from the vessel to a common project area on the National e-Infrastructure for Research Data (NIRD) (Sigma 2, 2024). For security, folders were backed up in NIRD and the Institute of Marine Research also held a copy of the data. Project members could apply for an account and access NIRD using secure shell or secure file transfer protocol (using software like WinSCP or FileZilla). Scientists were also encouraged to share their own Nansen Legacy data via the project area. This approach provided secure data access to project members only, whilst providing a shared working area for scientists to share and work on their data and prepare them for publication.
4.3 Training and support
The number of scientists actively using NIRD across Norway is growing. However, many project members were unfamiliar with using secure shell or secure file transfer protocol before the project. Dedicated project webinars and written training materials were provided to aid researchers in using NIRD.
4.4 Outcomes and Lessons Learned
Whilst many project members obtained user accounts to access the NIRD project area, data were instead often shared between project members via other methods. Furthermore, only a few scientists shared their own datasets with the rest of the project via NIRD. This was likely due to (1) analysis and quality control taking a long time for certain data (e.g. biological data), (2) the aforementioned unfamiliarity with using the NIRD platform for many project members, (3) reports that some project members were reluctant to share the data in their possession with other project members.
Although data were not always shared through NIRD, overall sharing increased during the project. Several practices that supported this are worth carrying forward. From the outset, we aimed to build trust and respect among scientists and institutions who might normally be in competition. Involving institutional data managers within the project proved particularly valuable, strengthening communication and collaboration both between data managers and scientists, and across institutions.
The metadata catalogue (Sect. 3.2.1) and the planned data publications document (Sect. 3.2.2) were useful in determining which datasets were not being shared and who was responsible for these datasets. However, governing this sensitive topic at scale within the project was deemed to be challenging and impractical and was instead managed on a case-by-case basis when data access was requested by a project member.
5.1 Motivation and Aims
The scientific community is growing increasingly aware of the importance of publishing FAIR data. The FAIR guiding principles aim to maximise the reuse of data, ensuring the greatest return on investment in terms of time, cost, and environmental impact involved in data collection. Central to the FAIR guiding principles is the requirement that data and metadata be fully readable and understandable by machines, a point emphasized throughout by Wilkinson et al. (2016). As the volume and heterogeneity of data continues to grow, the ability to automate the processing and integration of datasets becomes increasingly important. Big data presents both challenges and opportunities for data management and utilisation. Ensuring that data can be easily interpreted by machines is crucial for the development of services on top of data, such as:
-
Visualisation and analysis tools, allowing efficient and intuitive data exploration.
-
Streamlining the aggregation of multiple datasets into a single, usable file, providing flexibility and accessibility to the data users.
-
Options to download data into the user's choice of file format, ensuring flexibility and accessibility for diverse user needs.
Such services automate data preparation so that humans can focus on interpretation and analysis. These services are not merely technical conveniences; they provide the foundations for the creation of large scale, impactful projects that can serve humanity in significant ways. Despite the critical importance of standardised machine-readability, it is often overlooked in discussions about FAIR data, even by some online resources that discuss or provide guidance on publishing FAIR data.
The Nansen Legacy project's data management plan (The Nansen Legacy, 2024) emphasises that datasets should be published according to FAIR principles whenever possible. This approach maximises the value of the data for both the scientific community and society.
5.2 Methods
Nansen Legacy data should be published in machine-actionable data formats whenever possible (I, R) (The Nansen Legacy, 2024). Recommended formats are NetCDF files that adhere to the Climate and Forecast (CF) conventions (Eaton et al., 2024) and Darwin Core Archives (Darwin Core Community, 2010). All data should be findable through a data catalogue hosted by the Svalbard Integrated Arctic Earth Observing System (SIOS – https://sios-svalbard.org/metsis/search, last access: 6 November 2025), which aims to make all data relevant to Svalbard discoverable in one place (F). SIOS itself does not host any data; instead, the data catalogue harvests metadata from contributing data centers. As the Nansen Legacy is a Norwegian project, the following data centres were recommended (F, A):
-
Norwegian Marine Data Centre – https://metadata.nmdc.no/UserInterface/ (last access: 6 November 2025)
-
Norwegian Polar Data Centre – https://data.npolar.no/ (last access: 6 November 2025)
-
MET Arctic Data Centre – https://adc.met.no/ (last access: 6 November 2025)
-
NIRD Research Data Archive – https://archive.sigma2.no/ (last access: 6 November 2025)
However, a growing number of data centers hosted in other countries also contribute to SIOS, listed at https://sios-svalbard.org/DataSubmission, last access: 6 November 2025. These data centres have long-term commitments to storing, curating and make data available through time, including contingency plans for preservation if the service is shut down.
Data published to data centers that do not contribute to SIOS can be manually linked to the data catalog using a metadata collection form (https://sios-svalbard.org/metadata-collection-form, last access: 6 November 2025), though SIOS cannot build any services upon data linked using this approach.
5.3 Training and support
As part of the Nansen Legacy project, the following tools were developed to support data publication:
-
Nansen Legacy template generator. The spreadsheet template generator developed as part of Ellingsen et al. (2021) has been enhanced to help scientists both within and beyond the project prepare their metadata and data for publication in a structured manner (Marsden and Schneider, 2023). Users can select from the full list of CF standard names or Darwin Core terms to use as column headers. The templates include descriptions for each term as notes, appearing each time a cell is selected, and cell restrictions to prevent users from entering invalid values. The template generator includes configurations that facilitate the creation of Darwin Core Archives or CF-NetCDF files (I, R). This tool is being used and promoted outside of the project, including by SIOS, NorDataNet, OBIS, and the SCAR Antarctic Biodiversity Portal. The Nansen Legacy template generator is fully described by Marsden and Schneider (2024) and it is accessible at https://www.nordatanet.no/aen/template-generator/, last access: 6 November 2025.
-
Transforming Data from the Metadata Catalogue to Darwin Core event core and extensions. Project members recorded extensive metadata on each cruise, which is openly available in the metadata catalogue (Sect. 3.2.1). To avoid duplicating efforts by recording the same metadata again during data preparation for publication, a tool was developed to streamline this process. Scientists can provide the UUIDs related to their data records, and the tool returns spreadsheet templates pre-populated with associated metadata from the metadata catalogue. Each template includes multiple sheets that correspond to a core or extension in a Darwin Core Archive (I, R), including:
-
Event core – one row for each sampling event – https://rs.gbif.org/core/dwc_event_2024-02-19.xml (last access: 6 November 2025)
-
Occurrence extension – one row for each observation of an organism or group of organisms of the same species – https://rs.gbif.org/core/dwc_occurrence_2024-02-23.xml (last access: 6 November 2025)
-
Extended measurement or facts extension – one row for each measurement or fact related to either an event or occurrence – https://rs.gbif.org/extension/obis/extended_measurement_or_fact_2023-08-28.xml (last access: 6 November 2025)
It is relatively easy to create a Darwin Core Archive from the resulting product using the Integrated Publishing Toolkit (Robertson et al., 2014) developed by the Global Biodiversity Information Facility (GBIF). This process is described in a video tutorial at https://www.youtube.com/watch?v=ExtF2sSiH8s, last access: 6 November 2025, and the tool is hosted online at https://sios-svalbard.org/cgi-bin/aen_data/create_event_core_and_extensions.cgi, last access: 6 November 2025. Whilst this tool is tailored only to the Nansen Legacy metadata catalogue, we hope that this inspires developers in other projects that use metadata catalogues to develop similar tools to reduce the workload for their scientists.
-
Recognising the need for ongoing support and education, the Nansen Legacy project also provided training and resources to all project members. These resources were designed not only to teach the technical skills needed to publish FAIR data, but also to highlight the broader significance and impact of these practices. Some of these resources are available and applicable to the general scientific community beyond the project.
-
Presentations.
- –
Dedicated webinars were held to outline how to publish data in compliance with the project's data management plan.
- –
-
Workshops.
- –
Introductory workshops were held to teach researchers to work with CF-NetCDF files in Python or R. Attendees could create NetCDF files from dummy datasets and learn how to access data from real published datasets.
- –
Scientists were encouraged to bring their data and work on publishing them in either a Darwin Core Archive or CF-NetCDF files at dedicated workshops. Data managers were present to guide scientists through the process. These workshops were vital in helping scientists who are less familiar with creating such data formats.
- –
-
Video tutorials.
- –
One of the project's data managers, Luke Marsden, hosts a YouTube channel (https://www.youtube.com/@LukeDataManager, last access: 6 November 2025) where he shares video tutorials on how to work with FAIR data. This includes videos on how to create CF-NetCDF files in Python or R, how to extract data from CF-NetCDF files, and how to create Darwin Core Archives. Nansen Legacy has supported Luke in creating these videos.
- –
-
Written tutorials.
5.4 Outcomes and Lessons Learned
Our data management plan was ambitious, requiring significant changes in behaviour from many scientists. It is unsurprising that 100 % compliance was not achieved. However, the project has made a significant contribution to progressing the attitudes, habits and competence of its projects members which has likely had knock-on effects beyond the project. This is reflected in the following:
-
A growing number (hundreds) of Nansen Legacy datasets are accessible via the SIOS data catalogue. They can all be found in one place at https://sios-svalbard.org/metsis/search (last access: 6 November 2025) by filtering by collection using the abbreviation AeN (Arven etter Nansen), e.g https://sios-svalbard.org/metsis/search?f[0]=collection%3AAeN (last access: 6 November 2025).
-
The following datasets (amongst others) are published in CF-NetCDF files:
- -
CTD data (Reigstad et al., 2024)
- -
Nutrients data (Jones et al., 2024)
- -
- -
POC/PON data (e.g. Marquardt et al., 2022)
- -
Flow cytometry data (Müller et al., 2023)
- -
-
Biodiversity data and related measurements have been published in Darwin Core Archives, including data related to:
- -
Mesozooplankton (e.g. Wold et al., 2023)
- -
Phytoplankton (e.g. Assmy et al., 2022a)
- -
Ice algae (e.g. Assmy et al., 2022b)
- -
Sea ice meiofauna (e.g. Marquardt et al., 2023a)
- -
Publishing FAIR data is new for many scientists and there is a learning curve associated with this. There is therefore a clear need for better support and guidance. While some tools and frameworks have been developed – such as the FAIR Implementation Profile by the GO FAIR initiative (Magagna et al., 2020), which enables communities to articulate their approaches to FAIR data – there remains a significant need for additional tools and software to support and streamline all aspects of the FAIR data publishing workflow. Additionally, training resources should be made available to teach scientists how to publish and work with FAIR data effectively. It should not be overlooked, however, that making data FAIR can be both costly and challenging, particularly for smaller, heterogeneous datasets – often referred to as “long-tail data” – such as experimental results or diverse, novel field measurements collected by individual researchers or small teams. These datasets often require significant support to publish in a machine-actionable format. By clarifying the importance of publishing FAIR data and addressing these barriers, scientists will be more motivated and empowered to adopt these practices.
Despite the positive progress, we have identified several areas where data publishing practices could be improved to better support the FAIR principles. This section is divided into three subsections; the first focuses on challenges related to data centres, the second related to data formats, and the third related to granularity – a measure of how finely datasets are divided.
5.4.1 Data Centres
There is an ever-growing number of data centres. It is not practical for data users to have to search through all of these data centres to find data relevant to them. Data access portals aim to increase the findability (F) of data by making all data relevant to a certain region, or all data of a certain type, available in one place.
It is not practical for each data access portal to develop and maintain custom workflows to harvest metadata from each individual data centre. To build unified data access portals that truly expose all relevant data through a single access portal, data centres should host metadata systems that comply with commonly used standards (e.g. ISO 19115, GCMD DIF, EML, schema.org) and host their metadata on web platforms that consume this metadata system (e.g. OAI-PMH, OGC-CSW). This way, metadata harvesting workflows can be efficiently reused between many different data centres and data access portals (F).
Many popular data centres do not currently comply with commonly used standards. This reduces the Findability of data, thereby making reuse less likely. Like datasets, the accessibility and interoperability of data centres should also be considered when we discuss FAIR data (A, I).
5.4.2 Data Formats
The Nansen Legacy project has collected a wide range of datasets, presenting challenges in implementing the FAIR principles in some cases. While many of the Nansen Legacy datasets have been successfully published in machine-actionable data formats (I, R), there are instances where this has not been achieved. Throughout the project, we have gained valuable insights into the various reasons behind these shortcomings.
Firstly, as previously mentioned, there is a learning curve associated with working with FAIR data, particularly in creating and using machine-actionable data formats. It is evident that the data management community needs to provide greater support to scientists in this endeavour.
Secondly, it is not always obvious which data format scientists should choose for their data. Several measures can be taken to address this issue:
-
Greater availability of machine-actionable data formats. Suitable machine-actionable data formats do not exist for some complex scientific datasets. Existing data formats and conventions can be expanded to encompass more types of data where possible. However, while it may be necessary to develop new data formats and conventions, this should be approached with caution. Having fewer, broadly-used standards offers several advantages, such as more efficient development and maintenance, a smaller learning curve for data creators and users, and simplified data sharing between disciplines that use common data formats. Additionally, software and online tools that support access and visualisation of data can be developed and maintained more efficiently. Developing additional standards can be counterproductive, as it detracts from the goal of maintaining a limited set of standards to ensure consistency and interoperability.
-
Clarity on which data formats should be used. Guidance should be provided on what types of data should go into certain data formats. Examples should be included on how the data should be encoded.
-
A more proactive approach to developing standards. Most well-governed standards evolve in response to requests from the broader scientific or data management community. However, members of a scientific community who are not actively using a standard are unlikely to advocate for its development. A more proactive approach to expanding standards into new disciplines could therefore be valuable. It is unrealistic to expect scientists to dedicate significant time to mastering data standards such that they can adapt them to their needs. The data management community should play a key role in bridging this gap.
5.4.3 Granularity
Granularity is a measure of how finely datasets are divided, a crucial consideration for optimising data discovery and reuse. While data providers often group data by projects or research cruises for internal convenience or citation purposes, this approach can hinder data consumers who need aggregated datasets spanning regions or timeframes for numerical modelling, environmental monitoring, and other large-scale analyses. The Research Data Alliance Data Granularity Working Group (https://www.rd-alliance.org/groups/data-granularity-wg, last access: 6 November 2025) addresses these challenges by exploring solutions that balance the needs of both data providers and consumers. Fortunately, there are solutions to suit the needs of both data providers and consumers.
Publishing data with finer granularity provides several benefits:
-
Improved discoverability. Each dataset or profile is described with its own discovery and provenance metadata, making it easier for users to identify sampled locations and isolate the data they need (F, R).
-
Simplified dataset structure and enhanced workflows. Finer granularity reduces complexity by minimising the number of dimensions in individual datasets, which simplifies processing, interpretation, and integration into automated workflows or broader data networks (I).
-
Reduced redundancy in downloads. Users can download only the specific data they need, rather than larger aggregated datasets that may contain unnecessary information.
Common concerns about handling many small files – such as difficulties in downloading – can be addressed through improved data services. For example, data centres can provide tools to aggregate datasets upon user request. Data providers should recognise that users will increasingly be able to and interact with datasets in different formats and structures than those used for data storage. The focus should remain on creating datasets optimised for long-term storage and interoperability.
Some data centres support publishing data as collections, where each individual dataset is assigned its own metadata and DOI, and the collection as a whole also receives metadata and a DOI. This structure allows data users to cite either specific datasets or the entire collection, depending on the extent of data used, enhancing transparency in which data underpin publications. Journals could be encouraged to permit longer lists of references, enabling a greater number of datasets to be cited.
To maximise discoverability (F), reusability (R) and interoperability (I), we recommend the following best practices:
-
Publish at the highest functional granularity. Avoid combining data from multiple stations or sources into single datasets.
-
Separate datasets with different temporal resolutions. Minute-level and hourly-level observations, for instance, should not be merged.
-
Granularity in mixed-dimension datasets. Publish each feature type (vertical profile, trajectory, etc.) separately by default. Only combine when they share exactly the same coordinate axes and measurement context (e.g. a time-series of vertical profiles at a fixed location). Use explicit feature-type and dimension metadata – CF conventions' featureType is one example, but equivalent tags in other formats work just as well (Eaton et al., 2024).
-
Use metadata to establish relationships. Link datasets to research cruises, fieldwork, or other collection activities through tags or parent/child relationships, enabling discovery based on spatio-temporal criteria.
In the Nansen Legacy project, many have been advocating for finer granularity data, and several key data collections have been published in line with these recommendations (e.g. Vader, 2022; Müller et al., 2023).
No specific datasets were described or used in this article. Nansen Legacy data can be found via the SIOS data catalogue, filtering by collection using the abbreviation AeN (Arven etter Nansen), e.g. https://sios-svalbard.org/metsis/search?f[0]=collection%3AAeN (last access: 6 November 2025).
Effective data management in large-scale projects like the Nansen Legacy goes beyond technical systems and workflows; it also involves cultural and organisational shifts. These changes are essential for ensuring that data management practices are adopted, sustained, and continuously improved. A key to success in the Nansen Legacy project was integrating technical strategies with a strong emphasis on communication, coordination, and visibility of data management activites. The data management team were given the platform to provide oral presentations to all project members at each annual meeting within the project. Frequent email communications were sent to advertise data management webinars, share tutorials, and remind and encourage scientists to publish their data. The project's leadership, administration, and communication teams played a crucial role in echoing and amplifying the messages from the data management team. Positive feedback confirmed that it was helpful for project members to know they had a point of contact for their data management concerns.
A foundational aspect of effective data management was the establishment of a comprehensive data policy and data management plan at the project's inception. Our experiences underscored the need for these documents to be thorough, clear, and precisely worded. Misinterpretations of the open data policy sometimes led to incorrect assumptions about access to unpublished data. Additionally, it was not always clear which datasets were considered “Nansen Legacy data” and therefore needed to be managed adhering to the project's data policy and data management plan. This ambiguity was particularly challenging in cases where scientists were funded by multiple projects, or where external scientists participated in cruises funded by the Nansen Legacy project. This process will become easier if the adoption and enforcement of good data management practices become commonplace. In the meantime, agreeing on criteria for which datasets should comply with a project's data policy and data management plan at the project's inception would be beneficial.
Regular meetings were held involving data management representatives from all the research institutions participating in the project. This facilitated the relay of unified messages to each institution while also strengthening relationships between the institutions and affiliated data centres (Fig. 1). As discussed in Sect. 5.4.1, this kind of coordination is vital for building services that fully support FAIR data.
Effective data management extends beyond the confines of a single project and can be significantly supported by external drivers and incentives. For example, funding bodies can mandate that projects provide detailed data management plans and adhere to FAIR principles where feasible (e.g. Research Council of Norway, 2023). Additionally, the track record of implementing good data management practices should be considered alongside a scientist's paper publication record in funding and job applications. Making data management practices as efficient and user-friendly as possible is crucial, given the demanding schedules of scientists. Equally important is educating researchers on the significance of these practices and offering clear guidance on their implementation.
Key findings from the Nansen Legacy project include:
-
Consistent data collection. Sampling protocols developed collaboratively across the project enhanced consistency in data collection across the project. By adopting and further developing these protocols beyond the project, we can improve consistency in data collection across the scientific community. This would improve comparability of observations and enable more appropriate and accurate aggregation of datasets.
-
Keeping track of data collected. The logging system developed by Ellingsen et al. (2021) was widely adopted across the project, tracking data collected during research expeditions and making the metadata publicly available in a searchable online catalogue (https://sios-svalbard.org/aen/tools, last access: 6 November 2025). Future initiatives could focus on streamlining the process to reduce the workload for scientists.
-
Data storage and sharing. The National e-Infrastructure for Research Data (Sigma 2, 2024) hosted a centralised platform for storage and internal sharing of project data prior to publication. Adoption was uneven, most likely due to unfamiliarity with using secure file transfer protocol (SFTP) tools and reluctance to share data, often requiring case-by-case resolution. This highlights the need for training materials that not only advocate for best practices in data sharing and storage but also educate users on how to implement them.
-
Publishing FAIR data. The project mandated the publishing of FAIR data where possible, and provided tools and training that could be useful to scientists outside of the project (see Sect. 5.4.2). Despite progress, many scientists are still new to FAIR data publishing, indicating a need for further support, including the development of tools and software to streamline the process and training to ease the learning curve.
-
Implementation of policies. Successful implementation relied on more than just clear documentation. The cultural shift towards prioritising data management, coupled with consistent communication and support, played a crucial role. We recommend that data-management teams remain proactive in their communication and training, and that project leadership actively support and echo their messages. This support should include appointing dedicated data-management personnel to the project, allocating sufficient resources for training, infrastructure and coordination, and providing high-visibility platforms (e.g. plenary sessions, newsletters) for data-management updates. Leadership should also reiterate key data-management communications across governance meetings and partner institutions, and explicitly embed data-management milestones into project reporting.
In conclusion, our experiences from the Nansen Legacy project demonstrate that effective data management hinges on a blend of robust technical solutions and a supportive cultural environment. Success implementing the former hinges on the latter, and requires commitment from the project's leadership team. The experiences and practices developed through this project offer a valuable framework for future scientific endeavours, emphasising the need for continued focus on both technical and cultural aspects of data management.
LHM was the main data manager for the project from June 2020 until the end of the project (June 2024) and wrote most of the article. PGE was the main data manager for the project from May 2018 until August 2019, and has contributed to data management activities since. ØG and TMG were co-leads of the data management activity of the project. MR was the project leader. AM and HS helped manage the data flow of onboard instrumentation from the research vessels to the NIRD project area and published much of these data. HS, AM, ST, LF and ØG represented their data centres in the project data management group meetings and made significant contributions. TMG and MM helped in collecting an overview of project datasets to be published. MM coordinated the development of the sampling protocols documents. All authors have read the article and made contributions to the text.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors. Views expressed in the text are those of the authors and do not necessarily reflect the views of the publisher.
We would like to thank all the participants of the Nansen Legacy project who have used the metadata logging system or complied with the project's data policy and data management plan. Data collection and transfer from the vessels would not have been possible without the efforts of the crew onboard the vessels used (RV Kronprins Haakon, G.O. Sars, RV Kristine Bonnevie and briefly on MS Polarsyssel). Many thanks for the contributions of Benjamin Pfeil, Rahman Mankettikkara, Tomasz Kopec, Olaf Schneider, Rocio Castano Primo, Conrad Helgeland and Joël Durant who represented their institutions in the project data management group meetings. Dag Endresen, Rukaya Johaadien, Michal Torma and Vidar Bakken of GBIF Norway and Yi Ming Gan and Anton Van de Putte of the SCAR Antarctic Biodiversity Portal assisted the project in running workshops about Darwin Core. Magnar Martinsen helped to host the metadata catalogue on the SIOS website. Project data were stored in the NIRD project area NS9530K, NS9610K provided by Sigma2 – the National Infrastructure for High-Performance Computing and Data Storage in Norway, thanks to Maria Francesca Iozzi for supporting this.
This research has been supported by the Norges Forskningsråd (grant no. 276730).
This paper was edited by Kirsten Elger and reviewed by two anonymous referees.
Assmy, P., Gradinger, R., Edvardsen, B., Wiktor, J., Tatarek, A., Kubiszyn, A. M., Goraguer, L., and Wold, A.: Nansen Legacy JC2-1 phytoplankton biodiversity, https://doi.org/10.21334/npolar.2022.afe4302c [data set], sampling event dataset accessed via GBIF.org on 15 August 2024, 2022a. a
Assmy, P., Gradinger, R., Edvardsen, B., Wiktor, J., Tatarek, A., Smola, Z., Goraguer, L., and Wold, A.: Nansen Legacy JC2-1 ice algae biodiversity, https://doi.org/10.21334/npolar.2022.afe4302c [data set], sampling event dataset accessed via GBIF.org on 15 August 2024, 2022b. a
Darwin Core Community: Darwin Core: An Evolving Community-Developed Biodiversity Data Standard, http://www.tdwg.org/standards/450 (last access: 6 November 2025), version 1.4, 2010. a, b
Eaton, B., Gregory, J., Drach, B., Taylor, K., Hankin, S., Blower, J., Caron, J., Signell, R., Bentley, P., Rappa, G., Höck, H., Pamment, A., Juckes, M., Raspaud, M., Horne, R., Whiteaker, T., Blodgett, D., Zender, C., Lee, D., Hassell, D., Snow, A. D., Kölling, T., Allured, D., Jelenak, A., Sørensen, A. M., Gaultier, L., and Herlédan, S.: Climate and Forecast (CF) Metadata Conventions, https://cfconventions.org/ (last access: 6 November 2025), version 1.10, 2022. a
Eaton, B., Gregory, J., Drach, B., Taylor, K., Hankin, S., Blower, J., Caron, J., Signell, R., Bentley, P., Rappa, G., Höck, H., Pamment, A., Juckes, M., Raspaud, M., Horne, R., Whiteaker, T., Blodgett, D., Zender, C., Lee, D., Hassell, D., Snow, A. D., Kölling, T., Allured, D., Jelenak, A., Soerensen, A. M., Gaultier, L., Herlédan, S., Manzano, F., Bärring, L., Barker, C., and Bartholomew, S. L.: NetCDF Climate and Forecast (CF) Metadata Conventions, https://doi.org/10.5281/zenodo.11288138, 2024. a, b
Ellingsen, P. G., Ferrighi, L., Godøy, Ø. A., and Gabrielsen, T. M.: Keeping track of samples in multidisciplinary fieldwork, Data Science Journal, https://doi.org/10.5334/dsj-2021-034, 2021. a, b, c, d, e, f, g
Jones, E., Chierici, M., Lødemel, H. H., Møgster, J., Fonnes, L. L., and Fransson, A.: Water column data on dissolved inorganic nutrients (nitrite, nitrate, phosphate and silicic acid) from the Nansen Legacy cruises, https://doi.org/10.21335/NMDC-1698885798, 2024. a
Koenig, Z., Muilwijk, M., Sandven, H., Lundesgaard, Ø., Assmy, P., Lind, S., Assmann, K. M., Chierici, M., Fransson, A., Gerland, S., Jones, E., Renner, A. H., and Granskog, M. A.: From winter to late summer in the northwestern Barents Sea shelf: Impacts of seasonal progression of sea ice and upper ocean on nutrient and phytoplankton dynamics, Progress in Oceanography, 220, 103174, https://doi.org/10.1016/j.pocean.2023.103174, 2024. a
Magagna, B., Schultes, E. A., Pergl, R., Hettne, K. M., Kuhn, T., and Suchánek, M.: Reusable FAIR Implementation Profiles as Accelerators of FAIR Convergence, Springer International Publishing, Cham, 138–147, https://doi.org/10.31219/osf.io/2p85g, 2020. a
Marquardt, M., Patrohay, E., Goraguer, L., Dubourg, P., and Reigstad, M.: Concentration of Particulate Organic Carbon (POC) and Particulate Organic Nitrogen (PON) from the sea water and sea ice in the northern Barents Sea as part of the Nansen Legacy project, Cruise 2022702 JC3, [data set], https://doi.org/10.11582/2022.00052, 2022. a
Marquardt, M., Bluhm, B., and Gradinger, R.: Sea-ice meiofauna biodiversity from the Nansen Legacy cruise Q4 (cruise number: 2019711), https://doi.org/10.15468/gx9ujt [data set], sampling event dataset accessed via GBIF.org on 15 August 2024, 2023a. a
Marquardt, M., Goraguer, L., Assmy, P., Bluhm, B. A., Aaboe, S., Down, E., Patrohay, E., Edvardsen, B., Tatarek, A., Smoła, Z., Wiktor, J., and Gradinger, R.: Seasonal dynamics of sea-ice protist and meiofauna in the northwestern Barents Sea, Progress in Oceanography, 218, 103128, https://doi.org/10.1016/j.pocean.2023.103128, 2023b. a
Marsden, L.: How to publish FAIR Nansen Legacy data, https://doi.org/10.5281/zenodo.11067105, 2024a. a
Marsden, L.: NetCDF in Python – from beginner to pro, https://doi.org/10.5281/zenodo.10997447, 2024b. a
Marsden, L.: NetCDF in R – from beginner to pro, https://doi.org/10.5281/zenodo.11400754, 2024c. a
Marsden, L. and Schneider, O.: SIOS-Svalbard/Nansen_Legacy_template_generator: Nansen Legacy template generator, Zenodo [software], https://doi.org/10.5281/zenodo.8362212, 2023. a
Marsden, L. and Schneider, O.: The Nansen Legacy Template Generator for Darwin Core and CF-NetCDF, Data Science Journal, 23, https://doi.org/10.5334/dsj-2024-038, 2024. a
Müller, O., Petelenz, E., Tsagkaraki, T., Langvad, M., Olsen, L., Grytaas, A., Thiele, S., Stabell, H., Skjoldal, E., Våge, S., and Bratbak, G.: Flow cytometry measurements (abundance of virus, bacteria and small protists (primarily <20 µm)) during Nansen Legacy cruises, https://doi.org/10.21335/NMDC-1588963816 [data set], 2023. a, b
Reigstad, M., Fer, I., Ingvaldsen, R., Nilsen, F., Renner, A., Ludvigsen, M., Franson, A., Husum, K., Sundfjord, A., Gerland, S., Jones, E., Baumann, T., Søreide, J., Lundesgaard, Ø., and Husson, B.: CTD data from Nansen Legacy Cruises 2018–2022, https://doi.org/10.21335/NMDC-1174375695 [data set], 2024. a
Research Council of Norway: Sharing Research Data, https://www.forskningsradet.no/en/research-policy-strategy/open-science/research-data/, last access: 8 September 2023. a
Robertson, T., Döring, M., Guralnick, R., Bloom, D., Wieczorek, J., Braak, K., Otegui, J., Russell, L., and Desmet, P.: The GBIF integrated publishing toolkit: facilitating the efficient publishing of biodiversity data on the internet, PLOS ONE, 9, e102623, https://doi.org/10.1371/journal.pone.0102623, 2014. a
Sigma 2: National e-Infrastructure for Research Data Project Areas, Owned by Sigma 2 and operated by NRIS, https://documentation.sigma2.no/files_storage/nird_lmd.html (last access: 6 November 2025), 2024. a, b
Tenopir, C., Rice, N. M., Allard, S., Baird, L., Borycz, J., Christian, L., Grant, B., Olendorf, R., and Sandusky, R. J.: Data sharing, management, use, and reuse: Practices and perceptions of scientists worldwide, PLOS ONE, 15, e0229003, https://doi.org/10.1371/journal.pone.0229003, 2020. a, b
The Nansen Legacy: The Nansen Legacy Data Policy, https://doi.org/10.7557/nlrs.5799, 2021. a, b, c
The Nansen Legacy: Sampling Protocols: Version 10, https://doi.org/10.7557/nlrs.6684, 2022. a
The Nansen Legacy: Data Management Plan 2024, https://doi.org/10.7557/nlrs.7554, 2024. a, b, c, d, e, f
Vader, A.: Chlorophyll A and phaeopigments Nansen Legacy, https://doi.org/10.21335/NMDC-1371694848 [data set], 2022. a, b
Wassmann, P.: Chapter 3 – The Nansen Legacy: pioneering research beyond the present ice edge of the Arctic Ocean, in: Partnerships in Marine Research, edited by: Auad, G. and Wiese, F. K., Science of Sustainable Systems, Elsevier, 33–51, https://doi.org/10.1016/B978-0-323-90427-8.00009-5, 2022. a, b
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., et al.: The FAIR Guiding Principles for scientific data management and stewardship, Scientific Data, 3, 160018, https://doi.org/10.1038/sdata.2016.18, 2016. a, b, c
Wold, A., Søreide, J. E., Svensen, C., Halvorsen, E., Hop, H., Kwasniewski, S., and Ormańczyk, M.: Nansen Legacy JC1 mesozooplankton biodiversity, https://doi.org/10.21334/npolar.2022.f8d4a1cb [data set], sampling event dataset accessed via GBIF.org on 15 August 2024, 2023. a



