the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
The fortedata R package: open-science datasets from a manipulative experiment testing forest resilience
Jeff W. Atkins
Elizabeth Agee
Alexandra Barry
Kyla M. Dahlin
Kalyn Dorheim
Maxim S. Grigri
Lisa T. Haber
Laura J. Hickey
Aaron G. Kamoske
Kayla Mathes
Catherine McGuigan
Evan Paris
Stephanie C. Pennington
Carly Rodriguez
Autym Shafer
Alexey Shiklomanov
Jason Tallant
Christopher M. Gough
Ben Bond-Lamberty
The fortedata R package is an open data notebook from the Forest Resilience Threshold Experiment (FoRTE) – a modeling and manipulative field experiment that tests the effects of disturbance severity and disturbance type on carbon cycling dynamics in a temperate forest. Package data consist of measurements of carbon pools and fluxes and ancillary measurements to help analyze and interpret carbon cycling over time. Currently the package includes data and metadata from the first three FoRTE field seasons, serves as a central, updatable resource for the FoRTE project team, and is intended as a resource for external users over the course of the experiment and in perpetuity. Further, it supports all associated FoRTE publications, analyses, and modeling efforts. This increases efficiency, consistency, compatibility, and productivity while minimizing duplicated effort and error propagation that can arise as a function of a large, distributed and collaborative effort. More broadly, fortedata represents an innovative, collaborative way of approaching science that unites and expedites the delivery of complementary datasets to the broader scientific community, increasing transparency and reproducibility of taxpayer-funded science. The fortedata package is available via GitHub: https://github.com/FoRTExperiment/fortedata (last access: 19 February 2021), and detailed documentation on the access, used, and applications of fortedata are available at https://fortexperiment.github.io/fortedata/ (last access: 19 February 2021). The first public release, version 1.0.1 is also archived at https://doi.org/10.5281/zenodo.4399601 (Atkins et al., 2020b). All data products are also available outside of the package as .csv files: https://doi.org/10.6084/m9.figshare.13499148.v1 (Atkins et al., 2020c).
- Article
(1395 KB) - Full-text XML
-
Supplement
(295 KB) - BibTeX
- EndNote
Disturbance alters multiple carbon (C) cycling processes and, as a result, may affect forest C uptake and storage (Williams et al., 2016). The magnitude, timing, and duration of changes in the C cycle following disturbance vary among forests (Amiro et al., 2010; Luo and Weng, 2011; Coomes et al., 2012; Hicke et al., 2012; Gough et al., 2013; Peters et al., 2013; Vanderwel et al., 2013; Flower and Gonzalez-Meler, 2015; Gu et al., 2019). These responses may differ as a function of disturbance severity, type, and frequency along with the physical, structural, and biological properties of the affected ecosystem (Amiro et al., 2010; Williams et al., 2012; Scheuermann et al., 2018; Rebane et al., 2019; Fahey et al., 2020; Atkins et al., 2020a). Understanding which forest ecosystems are most vulnerable to disturbance and, conversely, what characteristics of an ecosystem confer C cycling stability remains an important frontier crucial to forecasting changes in the terrestrial C sink in the face of rising global disturbance frequencies (Frelich and Reich, 1999; White and Jentsch, 2001; Johnstone et al., 2010; 2016). Large-scale manipulative experiments may be particularly useful to identify the C fluxes and drivers that determine ecosystem C balance following disturbance (Fahey et al., 2020; Gough et al., 2013; Shiels and González, 2014).
Honing the prediction of how forests respond to disturbance, however, requires the parallel examination of mechanisms leading to the stability or decline of multiple C stocks and fluxes to changing disturbance regimes. The calculation and interpretation of forest ecosystem C balance necessitates repeated measurements of aboveground C stocks and fluxes through tree and litterfall inventories and belowground processes including root production and soil respiration – the total CO2 efflux from roots and microbes to the atmosphere. Complementary process and structural measurements such as leaf physiology, morphology, and chemistry along with remotely sensed measures of canopy structure and physiology provide important ancillary data useful to the interpretation of changes in C fluxes following disturbance. Few comprehensive datasets from such experiments exist in the public domain, and those that exist are almost never published in near-real time concurrently as an experiment is conducted, which limits testing hypotheses related to forest resilience and functional change beyond the focus of the project and slows the scientific enterprise more broadly (Falster et al., 2019).
The “open data” movement in science emphasizes transparency, reproducibility, and the moral imperative of making publicly funded research products broadly available (Culina et al., 2018). A specific example of this is “open notebook” science where the entire data record of a research project is made publicly available in near-real time with the goal of generating, integrating, documenting, and reporting heterogeneous data streams (e.g., Bond-Lamberty et al., 2016; Falster et al., 2019). Open notebook science helps create accountability and transparency by documenting the provenance of research data from conceptualization to publication and fights against the file drawer effect of lost data (Rosenthal, 1979). The ability for a project team to pull from one well-documented and consistent open data notebook increases research productivity and efficiency – streamlining the process of data curation and manipulation, and eliminating errors or inconsistencies that may otherwise be introduced from multiple copies of datasets across multiple workstations. In turn, this increases the potential for reproducibility and data use outside of a core project (Powers and Hampton, 2019; Schapira et al., 2019; Gallagher et al., 2020). Open data notebooks also perfectly complement mentoring and teaching – simultaneously serving to rapidly and effectively onboard new team members to the project while also providing project-based learning opportunities in the classroom that teach open science and data science skills.
The goal of this paper is to (i) describe the scientific context and goals of the Forest Resilience Threshold Experiment (FoRTE), (ii) describe its experimental design and high-level measurement protocols, and (iii) document the open-source fortedata package that serves as the project data repository. The systematically documented and transparent approach to science outlined in this paper and in the fortedata package surpasses the data-sharing expectations of publishers and funding bodies – specifically the publication of data prior to manuscript(s) submission – and may be considered as a model for future experiments and projects that is in line with widely adopted principles concerning the management and stewardship of scientific data (See FAIR Principles, Wilkinson et al., 2016).
FoRTE is a modeling and manipulative experiment that aims to identify the mechanisms underlying C cycling response to disturbance – specifically net primary productivity (NPP) resilience and its decline following disturbance. It centers on a manipulative field experiment located in northern lower Michigan at the University of Michigan Biological Station (45.58∘ N, 84.71∘ W) with experimental plots that span ∼ 8 ha of regionally representative landforms and forest types (Fig. 1). Data from the field experiment also inform a series of modeling experiments; specifically, data included in this package are used to initialize, calibrate, and validate dynamic vegetation model simulations of forest function and its responses to disturbance (e.g., Shiklomanov et al., 2021).
The experimental design follows a hierarchical structure with four replicates (A, B, C, D) of each factorial combination of disturbance severity (four levels) and type (two levels) (Fig. 1a, b). Within each replicate, each 0.5 ha plot was randomly assigned a disturbance severity level of 0 %, 45 %, 65 %, or 85 % gross defoliation, respectively (Fig. 1a). Each plot is bisected, with each half subjected to a disturbance treatment preferentially targeting large (top-down) or small (bottom-up) canopy trees (Fig. 1). All trees larger than 8 cm in diameter at breast height (DBH) are classified as canopy trees. An intensively surveyed 0.1 ha subplot is nested within each disturbance-severity–treatment combination – there are a total of 32 subplots (Fig. 1). The standard nomenclature for subplots is a concatenation of the replicate (A, B, C, D) plot number (01, 02, 03, 04) and subplot location (E for east side of the plot, or W for the west side of the plot) referred to in datasets by the variable name of subplot_id (Fig. 1b; Tables S2–S5 in the Supplement). Within each subplot, all canopy trees are measured (DBH) and geolocated (total no. of measured trees 3165; Fig. 2), and terrestrial laser scans using both 2D and 3D lidar (light detection and ranging) are taken to estimate canopy structural traits (Atkins et al., 2018; Fahey et al., 2019).
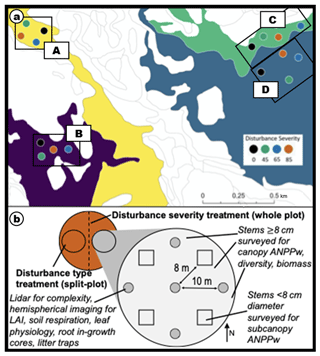
Figure 1(a) Map showing the distribution of plots in relation to landform types (Pearsall, 1996; Table S1). Plot replicates are grouped (A, B, C, D) with colors indicating severity level; (b) subplot diagram showing position of nested subplots for sampling and arrangement of subplots within the plot (orange).
Within each subplot, a series of C cycling and environmental measurements are taken at nested subplots. There are two types of nested subplot: (1) nested subplots 0, 1, 3, 5, and 7 are 1 m2 plots located at plot center (0) and 10 m off plot center at cardinal directions (1: north; 3: east; 5: south; and 7: west) (Fig. 1b), where environmental measurements such as soil volumetric water content, soil temperature, soil CO2 efflux, and hemispherical imagery are taken; (2) nested subplots 2, 4, 6, and 8 are 4 m2 vegetation survey plots located 8 m from plot center at intercardinal directions (2: northeast; 4: southeast; 6: southwest; and 8: northwest) (Fig. 1b), where understory leaf physiology, morphology, and chemistry measurements are taken. Additionally, all stems in the 4 m2 vegetation survey plots, including those below the 8 cm DBH canopy threshold, are counted and identified to the species level. The data detailed above are meant to be illustrative, but not entirely inclusive, of what is being measured in FoRTE. Additional environmental measurements will be taken as FoRTE matures and added to fortedata prior to incorporation in conventional data products such as research papers – including, but not limited to, soil chemical and physical properties, dendrometer readings, canopy profiles from 3D terrestrial lidar, fine root production, root density profiles, and data products from a NEON Airborne Observation Platform 2019 flyover. The fortedata readme file includes updates on the progress of current and future data availability.
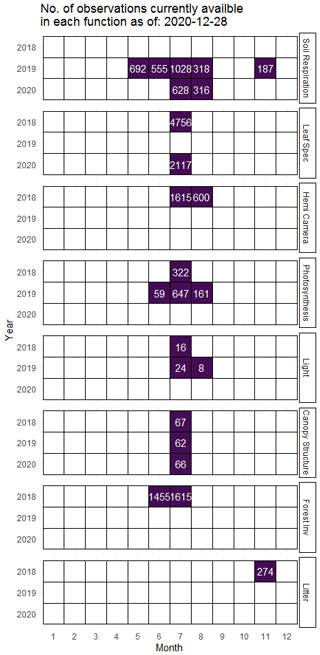
Figure 2Number of available records as of 13 December 2020 for time-series datasets including soil respiration (as well as soil temperature and soil water content) called from fd_soil_respiration; leaf spectrometry including leaf-level vegetation spectra indices from fd_spectrometry; hemispherical camera imagery including camera derived LAI, gap fraction, and NDVI from fd_hemi_camera; photosynthesis and stomatal conductance from fd_photosynthesis; light availability from fd_ceptometer; canopy structural traits from fd_canopy_structure; forest inventory data from fd_forest_inventory; and litter mass collected from litter traps from fd_litter.
2.1 The fortedata package
fortedata is a package for the R language (R Core Team, 2020) that includes field data from FoRTE. The fortedata package version 1.0.1 (Atkins et al., 2020b) includes leaf physiology, canopy structural traits, soil respiration, litterfall, soil micrometeorology, and forest inventory data for the years 2018, 2019, and 2020. Additional project data and data products will be incorporated over the lifetime of the project (initial FoRTE NSF funding 2018–2022).
2.2 Versioning and archiving
The fortedata package uses semantic versioning (https://semver.org/, last access: 19 February 2021), meaning version numbering follows an “x.y.z” format where x is the major version number, y the minor version number, and z is the patch version number. For example, this paper specifically details version 1.0.0. The major version number (x) only changes when there is a major change in overall package structure or there is expansive update in data – for example, following the inclusion of all data for a given field season. The minor version number (y) changes follow less notable changes, such as minor changes in functionality or the addition of minor data products. Changes in the patch version number (z) represent minor bug fixes or error corrections that do not affect package structure. Following each (major) release a DOI will be issued and the data archived by Zenodo (https://zenodo.org/, last access: 19 February 2021). All changes to data and code are immediately available through the GitHub repository, but only official releases will be issued a DOI.
2.3 Package license
The fortedata package is under a CC-BY-4 license (https://creativecommons.org/licenses/by/4.0/, last access: 19 February 2021); see the “LICENSE” file in the repository. This is identical to that used by, e.g., AmeriFlux and FLUXNET Tier 1. This license provides that users may copy and redistribute this R package and their associated data in any medium or format, adapting and building upon them for any scientific or commercial purpose, as long as appropriate credit is given. We request that users cite this paper (see Sect. 3.4) and strongly encourage them to (i) cite all constituent dataset primary publications (see fd_publications()) and (ii) involve data contributors as co-authors when possible and appropriate.
2.4 Citing the FoRTE data package
Papers or other research products using any FoRTE data should cite both this publication and the fortedata package, including the package version used. Appropriate citations can be found via the command citation(“fortedata”).
2.5 Using the FoRTE data package to access FoRTE data
It is necessary to install and use the fortedata R package in order to access FoRTE
data. The fortedata package can be installed directly from GitHub (https://github.com/FoRTExperiment/fortedata, last access: 19 February 2021) (Atkins et al., 2020b) using
the devtools package in R (Wickham et al., 2020):
devtools::install_github("FoRTExperiment /fortedata")
library(fortedata).
We plan to submit fortedata to the Comprehensive R Archive Network (CRAN), the common clearing house for all standardized R software packages.
2.6 FoRTE data package structure
The package is structured as a collection of independent datasets with standardized plot notation, date (ISO 8601 standard YYYY-MM-DD), and time (HH:MM:SS TZ) formatting (see fd_plot_metadata and Tables S1–S3 for more information). Datasets are available via user-facing, external functions outlined below. Additional metadata, instrument specifications, and abbreviated measurement protocols are available in the Supplement (Tables S1–S11) and in package documentation. Currently available functions include the following.
-
fd_inventory() returns a single dataset of the forest inventory data, including diameter at breast height (DBH), latitude, longitude, species, and information on vitality and canopy position (Fig. 3; Table S6). There are 3165 observations, all measured in 2018 (Figs. 2, 3). DBH measurements were taken with a Haglof PDII digital caliper (Haglof, Inc., Madison, MS, USA). Longitude and latitude were measured using a Trimble R1 GNSS receiver (Trimble; Sunnyvale, CA, USA), which has an accuracy range of ±30 cm. Re-measurement of DBH is slated for 2022. Additionally, mortality assignments per tree can be found in the fd_ mortality() function which is structurally similar to fd_inventory().
-
fd_soil_respiration() returns a single dataset currently with 3908 observations each of soil CO2 efflux (), soil temperature (∘C; integrated from 0 to 7 cm depth), and volumetric water content (%) for the years 2019 and 2020 (Figs. 2, 4; Table S7). Soil CO2 efflux was measured using a LI-6400 XT (LI-COR Biosciences; Lincoln, NE) with a soil CO2 flux chamber model 6400-09 attachment with a measurement accuracy of ±5 µmol mol−1 maximum deviation. Soil temperature was measured using the attached soil temperature probe, with an accuracy of ±1.5 ∘C. Soil moisture was measured using a Campbell HS2 HydroSense II time domain reflectometer (Campbell Scientific; Logan, UT, USA) with a measurement accuracy of ±3 % and accurate range of 0 %–50 %.
-
fd_leaf_spectrometry() returns a single dataset of vegetation indices derived from leaf-level spectrometry data collected via a CI-710 handheld spectrometer (Table S8). The dataset currently includes 6873 observations from 2018 and 2020 of spectral indices for three species each in eight subplots within the D replicate (Figs. 1 and 2).
-
fd_photosynthesis() returns a single dataset of leaf physiology variables, including photosynthesis and transpiration measured using a LI-6400 XT (LI-COR Biosciences; Lincoln, NE) (Table S9) with a measurement accuracy of ±5 µmol mol−1 maximum deviation. The dataset includes 2215 observations from 2018 (Fig. 2).
-
fd_litter() returns a single dataset of litter mass collected via litter traps (four in each subplot, at nested sampling points 1, 3, 5, 7). The data include the tare + oven-dried mass for each litter fraction as well as the tare weight (the empty bag), by subplot (Fig. 5; Table S10). The data are coded by litter fraction, denoted in the fraction column as either leaf, fwd (fine woody debris), or misc (miscellaneous, unidentifiable leaf fragments). Litter mass can be calculated by subtracting the tare weight from the mass + tare. There are a total of 340 observations included in the dataset from 2018, with 2019 and 2020 data to be processed and added in early 2021 (Fig. 5).
-
fd_hemi_camera() returns a single dataset that includes derived estimates of leaf area index, gap fraction, clumping index, and NDVI (normalized difference vegetation index) from terrestrial, upward-facing hemispherical photos looking into the forest canopy taken 1 m above ground (Table S11). The dataset includes 1028 observations of each variable from 2018 and 2019 (Fig. 2).
-
fd_canopy_structure() returns a single dataset that includes estimates of canopy structural traits such as height, area/density, openness, complexity, and arrangement derived from terrestrial lidar and processed using forestr version 1.0.1 (Atkins et al., 2018) in R version 3.6.2 (R Core Team, 2020). The package includes 195 observations for each metric (28 canopy structural metrics are included in forestr v1.0.1 that estimate canopy structural traits such as area/density, openness, arrangement, heterogeneity, and layering (Atkins et al., 2018; Fahey et al., 2019)) from 2018, 2019, and 2020 (Table S12).
-
fd_ceptometer() returns a single dataset that includes estimates of the fraction of photosynthetically available radiation (faPAR) absorbed by the canopy as well as leaf area index (LAI) – each derived from a handheld ceptometer (LP-80; Decagon Devices) (Table S13) with a resolution of 1 and accuracy of ±5 %. The dataset includes 32 observations of each variable from 2019 and 16 from 2018 (Fig. 2).
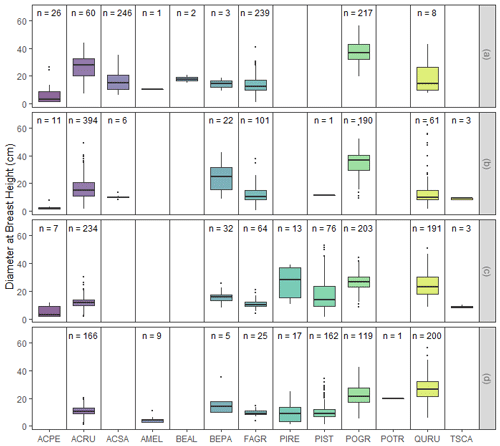
Figure 3Diameter at breast height (DBH) distributions for each species, grouped by replicate. The bounds of each box in the box plot represent the 25th percentile at the lower bound and the 75th percentile at the upper bound, and the horizontal line is the median. Lines extending from the lower and upper bounds represent values that are 1.5 times the interquartile range for the minimum and maximum values, respectively, while black circles indicate outliers. Above each box plot, n is the number of observations.
Additionally, fortedata includes functionality beyond simple data ingestion.
-
Brief summaries of certain datasets are available via summary functions, such as fd_inventory_summary(), which returns a summary of the fd_inventory() dataset that includes stocking density (in stems ha−1) and mean basal area (m2 ha−1) averaged at the subplot level (n=32) grouped by replicate, plot, and subplot variables. fd_canopy structure_summary() returns a similar table of canopy structural trait data.
-
Experimental design information, including plot metadata such as disturbance severity or treatment assignments, can be accessed via fd_plot_metadata() (see FoRTE Working with Data vignette for a worked example).
-
Biomass estimates from plot forest inventory data are available using the calc_biomass() function, which uses regionally relevant allometries with a power law function to convert tree diameter to biomass in kilograms of C and calc_lai() estimates leaf area index (LAI) from FoRTE litter data found in fd_litter() using site-derived specific leaf area (SLA) data (see Leaf Litter vignette for further information).
2.7 Accessing FoRTE data without using fortedata
All data contained in fortedata can also be accessed directly via Figshare (https://doi.org/10.6084/m9.figshare.12292490.v3, Atkins et al., 2020c) as a compressed file containing all output generated from each function in fortedata (Atkins et al., 2020c). This mirror of the dataset will be updated with each major release of fortedata.
2.8 FoRTE documentation and vignettes
This paper serves as the primary documentation for FoRTE data, and all code to reproduce this paper – including the tables and plots herein – is available in the package (https://github.com/FoRTExperiment/fortedata/tree/master/essd, last access: 19 February 2021). The package also includes additional supporting documentation via R's standard help system. Vignettes, which are guided tutorials that include example code or background information such as experimental design and proposal narratives, are also included both in the package and online and can be accessed via BrowseVignettes(“fortedata”). Vignettes are currently available for the functions above, and additional vignettes will be added as new data products are incorporated into fortedata.
Supporting project information, including detailed methods and data collection information (introduced briefly below and in Supplement Tables S1–S11), can be found within package documentation: function help files (e.g., ?fd_inventory()) and package vignettes – which can be accessed via browseVignettes(“fortedata”) or online at https://fortexperiment.github.io/fortedata/ (last access: 19 February 2021). The funded project narrative (NSF DEB-165509) can be accessed directly within in the package via vignette(“fd_forte_ proposal_ vignette”) and outlines hypotheses, objectives, proposed methods, and supporting literature for the project.
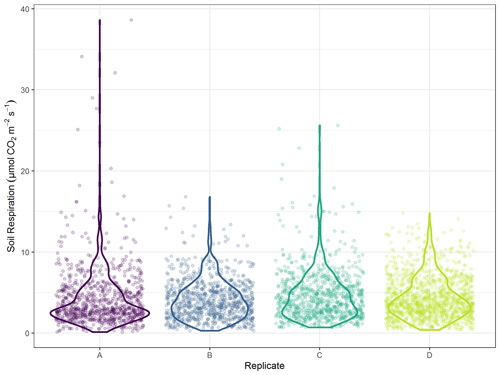
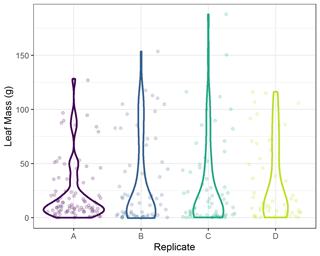
Figure 4Distribution of soil CO2 efflux values from May–November 2019 by replicate. Lines represent distribution, while points are individual measures.
2.9 Testing and quality assurance
The fortedata R package has a wide variety of unit tests that test code functionality, typically via assertions about function behavior, but also by verifying behavior of those functions when importing datasets. As datasets within fortedata differ in composition and format, they may create a variety of errors. Unit tests, detailed below, ensure that entries in these datasets are realistic and valid. These tests are run automatically every time fortedata code or data are updated on GitHub, ensuring continuing package validity for end users. These tests include error checks on the following:
-
appropriate date and timestamp formatting
-
data class verification (e.g., plot numbers as integer values, soil CO2 efflux measurements as numeric values)
-
out-of-bound latitude or longitude values
-
appropriately formatted plot metadata that adheres to FoRTE naming conventions
-
out-of-bound values (e.g., unreasonable, unrealistic, erroneous entries) for environmental measurements (e.g., negative values for tree DBH, soil water content < 0 or > 100).
The appropriate method of uncertainty quantification for any given dataset, herein fortedata, may vary based on the use, application, or analyses of these data. To this end, we have provided extensive documentation for end users to make these calculations based on their own judgement, discretion, or discipline-specific needs. This is why there is no direct quantification of uncertainty for datasets contained within fortedata. These data are raw and represent unmodified point measurements, taken according to each instrument's or method's standards. Any uncertainties associated with measurements, either instrument or method specific, are detailed above in Sect. 4.2 and in Tables S5–S11.
2.10 Reporting issues
We use the fortedata GitHub issue tracker (https://github.com/FoRTExperiment/fortedata/issues, last access: 19 February 2021) to track and categorize user improvement suggestions, problems, or errors with the R package code and included data, as well as requests for new variables or functionality, and/or other questions. All past and current issues are viewable to the public, and new issues can be contributed by anyone with a (free) GitHub account.
fortedata is available via GitHub (https://github.com/FoRTExperiment/fortedata, last access: 19 February 2021) and can be installed and accessed directly within the R programming language as outlined above. Additionally, the first version of fortedata (version 1.0.1) outlined in this paper is archived at https://doi.org/10.5281/zenodo.3936146 (Atkin et al., 2020b). We have also made all package data products accessible as formatted .csv files with accompanying documentation available via Figshare: https://doi.org/10.6084/m9.figshare.12292490.v3 (Atkins et al., 2020c).
The lack of existing publicly available datasets comprehensively documenting forest and ecosystem manipulations limits our ability to test hypotheses related to forest resilience and functional change, broadly. While projects such as FoRTE push our boundaries of understanding the mechanisms that facilitate ecological resilience, the additional effort to make the project as open and transparent as possible, including the expeditious delivery of project data, increases the impact of the project. FoRTE and the fortedata package serve as one model for future experiments and projects by showcasing the advantages of supplying centralized project data openly and to investigators within and external to the project. This approach is above and beyond the typical requirements and expectations for data availability, particularly in field-based ecology where standard conventions for data availability, if and where they do exist, call for reporting only upon project completion or publication. The results of such modular practices often limit data availability to single spreadsheets of varying quality with limited, sometimes non-existent, metadata. We argue that open-notebook science should be the new science normal, whenever possible – when we fail to provide timely, open, and usable data, we fall short of our duty as scientists and in doing so jeopardize scientific advancement and its societal benefits:
the free, open, and responsible practice of science is fundamental to scientific advancement for both human and environmental well-being. Science requires freedom of movement, collaboration, and communication, as well as equitable access to data and resources. It requires scientists to conduct and communicate scientific work for the benefit of society, with excellence, integrity, respect, fairness, trustworthiness, clarity, and transparency. (American Geophysical Union, 2017)
We do acknowledge there may be legitimate barriers for some scientists/project teams – such as limited access to reliable internet, to resources to acquire necessary computational skills, to budgeted time, or to supportive and collaborative environments where open-science is rewarded – these challenges require our attention and support. In addition, some types of data (proprietary, human subject) clearly require different standards and practices. That said, where there exists the privilege of having access to the necessary resources to conduct science openly and equitably, choosing to do otherwise is unconscionable. Open science approaches should be the rule and not the exception, and we anticipate that the release of fortedata in near-real time will motivate external collaboration, facilitate data exchange within the project, and provide project-wide data transparency, consistency, and availability, as well as increased team member efficiency and productivity.
The supplement related to this article is available online at: https://doi.org/10.5194/essd-13-943-2021-supplement.
JWA, BBL, and CMG wrote the manuscript. JWA, EAA, AB, KMD, LTH, LJH, MSG, AGK, KM, CM, EP, CR, AS, and JT collected, processed, and provisioned data for the package, including providing details on methods and metadata included within the manuscript and in package documentation. JWA, BBL, KD, SCP, and AS contributed code and package oversight. BBL and CMG envisioned, proposed, and oversaw the project. All authors contributed to manuscript discussion, editing, and revision.
The authors declare that they have no conflict of interest.
FoRTE is funded by the National Science Foundation (grant no. DEB-1655095). NEON AOP data collection was supported by NSF award no. 1702379 to Kyla M. Dahlin. Carly Rodriguez, Laura J. Hickey, and Evan Paris, in whole or in part, were supported by the UMBS REU program (NSF no. 1659338). We would also like the gratefully acknowledge the University of Michigan Biological Station for their continued support.
This research has been supported by the NSF (grant nos. DEB-1655095, 1702379, and 1659338).
This paper was edited by David Carlson and reviewed by Joseph Stachelek and one anonymous referee.
American Geophysical Union: The Responsibilities and Rights of Scientists, available at: https://www.agu.org/Share-and-Advocate/Share/Policymakers/Position-Statements/Rights-and-responsibilities-of-scientists (last access: 23 April 2020), 2017.
Amiro, B. D., Barr, A. G., Barr, J. G., Black, T. A., Bracho, R., Brown, M., Chen, J. M., Clark, K. L., Davis, K. J., Desai, A. R., Dore, S., Engel, V., Fuentes, J. D., Goldstein, A. H., Goulden, M. L., Kolb, T. E., Lavigne, M. B., Law, B. E., Margolis, H. A., Martin, T. A., McCaughey, J. H., Misson, L., Montes-Helu, M., Noormets, A., Randerson, J. T., Starr, and G. Xiao, J.: Ecosystem carbon dioxide fluxes after disturbance in forests of North America, J. Geophys. Res.-Biogeo., 115, G00K02, https://doi.org/10.1029/2010JG001390, 2010.
Atkins, J. W., Bohrer, G., Fahey, R. T., Hardiman, B. S., Morin, T. H., Stovall, A. E., and Gough, C. M.: Quantifying vegetation and canopy structural complexity from terrestrial LiDAR data using the forestr r package, Methods Ecol. Evol., 9, 2057–2066, 2018.
Atkins, J. W., Bond-Lamberty, B., Fahey, R. T., Hardiman, B. S., Haber, L., Stuart-Haëntjens, E., and Tallant, J.: Multidimensional Structural Characterization is Required to Detect and Differentiate Among Moderate Disturbance Agents, Ecosphere, 11, 1–19, https://doi.org/10.1002/ecs2.3156, 2020a.
Atkins, J. W., Bond-Lamberty, B., Dorheim, K., Pennington, S., and Shiklomanov, A.: fortedata v1.0.2 (Version 1.0.2), Zenodo, https://doi.org/10.5281/zenodo.4399601, 2020b.
Atkins, J. W., Bond-lamberty, B., Dorheim, K., Pennington, S. C., Shiklomanov, A., Agee, E., Gough, C. M., Shiklomanov, A., Dorheim, K., Pennington, S., Barry, A., Dahlin, K., Grigri, M., Haber, L., Hickey, L., Kamoske, A., Mathes, K., McGuigan, C., Paris, E., Rodriguez, C., Shafer, A., and Tallant, J.: fortedata-1.0.2, Dataset, figshare, https://doi.org/10.6084/m9.figshare.13499148.v1, 2020c.
Bond-Lamberty, B., Smith, A. P., and Bailey, V.: Running an open experiment: transparency and reproducibility in soil and ecosystem science, Environ. Res. Lett., 11, 084004, https://doi.org/10.1088/1748-9326/11/8/084004. 2016.
Coomes, D. A., Holdaway, R. J., Kobe, R. K., Lines, E. R., and Allen, R. B.: A general integrative framework for modelling woody biomass production and carbon sequestration rates in forests, J. Ecol., 100, 42–64, 2012.
Culina, A., Baglioni, M., Crowther, T. W., Visser, M. E., Woutersen-Windhouwer, S., and Manghi, P.: Navigating the unfolding open data landscape in ecology and evolution, Nature E and E, 2, 420–426, 2018.
Fahey, R. T., Atkins, J. W., Gough, C. M., Hardiman, B. S., Nave, L. E., Tallant, J. M., and Haber, L. T.: Defining a spectrum of integrative trait-based vegetation canopy structural types, Ecol. Lett., 22, 2049–2059, 2019.
Fahey, R. T., Atkins, J. W., Campbell, J. L., Rustad, L. E., Duffy, M., Driscoll, C. T., Fahey, T. J., and Schaberg, P. G.: Effects of an experimental ice storm on forest canopy structure, Can. J. Forest Res., 50, 136–145, 2020.
Falster, D. S., FitzJohn, R. G., Pennell, M. W., and Cornwell, W. K.: Datastorr: a workflow and package for delivering successive versions of “evolving data” directly into R, GigaScience, 8, giz035, https://doi.org/10.1093/gigascience/giz035, 2019.
Flower, C. E. and Gonzalez-Meler, M. A.: Responses of temperate forest productivity to insect and pathogen disturbances, Annu. Rev. Plant Biol., 66, 547–569, 2015.
Frelich, L. E. and Reich, P. B.: Minireviews: Neighborhood Effects, Disturbance Severity, and Community Stability in Forests, Ecosystems, 2, 151–166, 1999.
Gallagher, R. V., Falster, D. S., Maitner, B. S., Salguero-Gómez, R., Vandvik, V., Pearse, W. D., and Ankenbrand, M. J.: Open Science principles for accelerating trait-based science across the Tree of Life, Nature Ecology & Evolution, 4, 294–303, 2020.
Gough, C. M., Hardiman, B. S., Nave, L. E., Bohrer, G., Maurer, K. D., Vogel, C. S., Nadelhoffer, K. J., and Curtis, P. S.: Sustained carbon uptake and storage following moderate disturbance in a Great Lakes forest, Ecol. Appl., 23, 1202–1215, 2013.
Gough, C. M., Atkins, J. W., Bond-Lamberty, B., Agee, E. A., Dorheim, K. R., Fahey, R. T., Grigri, M. S., Haber, L. T., Mathes, K. C. Pennington, S. C., Shiklomanov, A. N., and Tallant, J. M.: Forest Structural Complexity and Biomass Predict First-Year Carbon Cycling Responses to Disturbancem Ecosystems, 1–14, https://doi.org/10.1007/s10021-020-00544-1, 2020.
Gu, H., Williams, C. A., Hasler, N., and Zhou, Y.: The carbon balance of the southeastern US forest sector as driven by recent disturbance trends, J. Geophys. Res.-Biogeo., 124, 2786–2803, 2019.
Hicke, J. A., Allen, C. D., Desai, A. R., Dietze, M. C., Hall, R. J., Hogg, E. H., and Vogelmann, J.: Effects of biotic disturbances on forest carbon cycling in the United States and Canada, Glob. Change Biol., 18, 7–34, 2012.
Johnstone, J. F., McIntire, E. J., Pedersen, E. J., King, G., and Pisaric, M. J.: A sensitive slope: estimating landscape patterns of forest resilience in a changing climate, Ecosphere, 1, 1–21, 2010.
Johnstone, J. F., Allen, C. D., Franklin, J. F., Frelich, L. E., Harvey, B. J., Higuera, P. E., and Schoennagel, T.: Changing disturbance regimes, ecological memory, and forest resilience, Front. Ecol. Environ., 14, 369–378, 2016.
Luo, Y. and Weng, E.: Dynamic disequilibrium of the terrestrial carbon cycle under global change, Trends Ecol. Evol., 26, 96–104, 2010.
Pearsall, D. R.: Landscape ecosystems of the University of Michigan Biological Station: Ecosystem diversity and ground-cover diversity, Doctoral dissertation, The University of Michigan, 1996.
Peters, E. B., Wythers, K. R., Bradford, J. B., and Reich, P. B.: Influence of disturbance on temperate forest productivity, Ecosystems, 16, 95–110, 2013.
Powers, S. M. and Hampton, S. E.: Open science, reproducibility, and transparency in ecology, Ecol. Appl., 29, e01822, https://doi.org/10.1002/eap.1822, 2019.
Rebane, S., Jõgiste, K., Põldveer, E., Stanturf, J. A., and Metslaid, M.: Direct measurements of carbon exchange at forest disturbance sites: a review of results with the eddy covariance method, Scand. J. Forest Res., 34, 585–597, 2019.
Rosenthal, R.: The file drawer problem and tolerance for null results, Psychological Bulletin, 86, 638–641, 1979.
Schapira, M., Harding, R. J., and The Open Lab Notebook Consortium: Open laboratory notebooks: good for science, good for society, good for scientists, F1000Research, 8, 87, https://doi.org/10.12688/f1000research.17710.1, 2019.
Scheuermann, C. M., Nave, L. E., Fahey, R. T., Nadelhoffer, K. J., and Gough, C. M.: Effects of canopy structure and species diversity on primary production in upper Great Lakes forests, Oecologia, 2, 405–415, https://doi.org/10.1007/s00442-018-4236-x, 2018.
Shiels, A. B. and González, G.: Understanding the key mechanisms of tropical forest responses to canopy loss and biomass deposition from experimental hurricane effects, Forest Ecol. Manage., 332, 1–10, 2014.
Shiklomanov, A. N., Bond-Lamberty, B., Atkins, J., and Gough, C. M.: Structure and parameter uncertinty in centennial projections of forest community structure and carbon cycling, Glob. Change Biol., 26, 6080–6096, https://doi.org/10.1111/gcb.15164, 2021.
Vanderwel, M. C., Coomes, D. A., and Purves, D. W.: Quantifying variation in forest disturbance, and its effects on aboveground biomass dynamics, across the eastern United States, Glob. Change Biol., 19, 1504–1517, 2013.
White, P. S. and Jentsch, A.: The Search for Generality in Studies of Disturbance and Ecosystem Dynamics, in: Progress in Botany: Genetics Physiology Systematics Ecology, edited by: Esser, K., Lüttge, U., Kadereit, J. W., and Beyschlag, W., Springer Berlin Heidelberg, Berlin, Heidelberg, 399–450, 2001.
Wickham, H., Hester, J., and Change, W.: devtools: Tools to Make Developing R Packages Easier, R package version 2.3.0, available at: https://CRAN.R-project.org/package=devtools (last access: 15 January 2021), 2020.
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., Blomberg, N., Boiten, J. W., da Silva Santos, L. B., Bourne, P. E., and Bouwman, J.: The FAIR Guiding Principles for scientific data management and stewardship, Sci. Data, 3, 1–9, https://doi.org/10.1038/sdata.2016.18, 2016.
Williams, C. A., Collatz, G. J., Masek, J. G., and Goward, S. N.: Carbon consequences of forest disturbance and recovery across the conterminous United States, Global Biochem. Cy., 26, GB1005, https://doi.org/10.1029/2010GB003947, 2012.
Williams, C. A., Gu, H., MacLean, R., Masek, J. G., and Collatz, G. J.: Disturbance and the carbon balance of US forests: A quantitative review of impacts from harvests, fires, insects, and droughts, Global Planet. Change, 143, 66–80, 2016.