the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
PhytoBase: A global synthesis of open-ocean phytoplankton occurrences
Damiano Righetti
Meike Vogt
Niklaus E. Zimmermann
Michael D. Guiry
Nicolas Gruber
Marine phytoplankton are responsible for half of the global net primary production and perform multiple other ecological functions and services of the global ocean. These photosynthetic organisms comprise more than 4300 marine species, but their biogeographic patterns and the resulting species diversity are poorly known, mostly owing to severe data limitations. Here, we compile, synthesize, and harmonize marine phytoplankton occurrence records from the two largest biological occurrence archives (Ocean Biogeographic Information System, OBIS; and Global Biodiversity Information Facility, GBIF) and three independent recent data collections. We bring together over 1.36 million phytoplankton occurrence records (1.28 million at the level of species) for a total of 1704 species, spanning the principal groups of the diatoms, dinoflagellates, and haptophytes, as well as several other groups. This data compilation increases the amount of marine phytoplankton records available through the single largest contributing archive (OBIS) by 65 %. Data span all ocean basins, latitudes, and most seasons. Analyzing the oceanic inventory of sampled phytoplankton species richness at the broadest spatial scales possible using a resampling procedure, we find that richness tends to saturate at ∼93 % of all species in our database in the pantropics, at ∼64 % in temperate waters, and at ∼35 % in the cold Northern Hemisphere, while the Southern Hemisphere remains under-explored. We provide metadata on the cruise, research institution, depth, and date for each data record, and we include phytoplankton cell counts for 193 763 records. We strongly recommend consideration of spatiotemporal biases in sampling intensity and varying taxonomic sampling scopes between research cruises or institutions when analyzing the occurrence data spatially. Including such information into predictive tools, such as statistical species distribution models, may serve to project the diversity, niches, and distribution of species in the contemporary and future ocean, opening the door for quantitative macroecological analyses of phytoplankton. PhytoBase can be downloaded from PANGAEA: https://doi.org/10.1594/PANGAEA.904397 (Righetti et al., 2019a).
- Article
(8363 KB) - Full-text XML
- BibTeX
- EndNote
Phytoplankton are photosynthetic members of the plankton realm, responsible for about half of the global net primary production (Field et al., 1998). While more than 4300 phytoplankton species have been described (Sournia et al., 1991), spanning at least six major clades (Falkowski et al., 2004), there are likely many more species living in the ocean, perhaps more than 10 000 (de Vargas et al., 2015). Some of these species (e.g., Emiliania huxleyi, Gephyrocapsa oceanica) are abundant and occur throughout the ocean (Iglesias-Rodríguez et al., 2002), but a majority of plankton species form low-abundance populations (Ser-Giacomi et al., 2018) and remain essentially uncharted; i.e., the quantitative description of where they live and where they do not is rather poor. This biogeographic knowledge gap stems from a lack of systematic global surveys that are similar to those undertaken for inorganic carbon (WOCE/JGOFS/GOSHIP; Wallace, 2001) or trace metals (GEOTRACES; Mawji et al., 2015). Owing to logistical and financial challenges associated with internationally coordinated surveys, our knowledge of phytoplankton biogeography is, with a few exceptions (Bork et al., 2015; McQuatters-Gollop et al., 2015), mostly based on spatially very limited surveys or basin-scale studies (e.g., Endo et al., 2018; Honjo and Okada, 1974). Marine phytoplankton occurrence data are unevenly distributed, incomplete in remote areas, and orders of magnitude higher in more easily accessed areas, especially near coasts (Buitenhuis et al., 2013). Additional factors that have impeded progress in developing a good biogeographic understanding of phytoplankton are difficulties in species identification, linked to their microscopic body size. This is reflected well in the current geographic knowledge of phytoplankton species richness from direct observations (e.g., Rodríguez-Ramos et al., 2015), which is much more limited compared to that of other marine taxa, such as zooplankton (e.g., Rombouts et al., 2010), fishes (e.g, Jones and Cheung, 2015), sharks (e.g., Worm et al., 2005), or krill (e.g., Tittensor et al., 2010), even though many of these taxa also suffer from deficiencies in sampling efforts (Menegotto and Rangel, 2018).
Initial efforts to overcome the data sparseness and patchiness for phytoplankton by the MareDat project (Buitenhuis et al., 2012; Leblanc et al., 2012; Luo et al., 2012; O'Brien et al., 2013; Vogt et al., 2012) resulted in the compilation and synthesis of 119 phytoplankton species from 17 240 sampling events. While representing a large step forward, the coverage remained relatively limited, largely owing to MareDat's focus on abundance data, motivated by the need to use the data for model evaluation and other quantitative assessments (Buitenhuis et al., 2013). However, during these efforts, it became clear that there are at least an order of magnitude more data in archives around the world if one relaxes the abundance criterion and considers all observations that included presences. The potential for the use of presences to constrain, e.g., phytoplankton community structure and richness, is large, as demonstrated by Righetti et al. (2019b), who recently produced the first global map of phytoplankton species richness. This application was also made possible thanks to the rapid developments in data-mining and statistical analysis tools, such as species distribution models (SDMs) (Guisan and Zimmermann, 2000) that permit scientists to account for some of the limitations stemming from the spatiotemporal sampling biases underlying species' occurrence data (Breiner et al., 2015; Phillips et al., 2009).
A key enabler for the compilation and synthesis of phytoplankton occurrences (presence or abundance records) is the existence of two digital biological data archives, i.e., the Global Biodiversity Information Facility (GBIF; https://www.gbif.org/, last access: 27 February 2017) and the Ocean Biogeographic Information System (OBIS; https://www.obis.org/, last access: 6 March 2017). GBIF is the world's largest archive for species occurrence records, while OBIS is the largest occurrence database on marine taxa. Both archives have gathered a large number of phytoplankton occurrence records and make them freely available to the global community. In addition to MareDat (Buitenhuis et al., 2013), marine surveys such as those conducted with the Continuous Plankton Recorder (CPR) (McQuatters-Gollop et al., 2015), the Atlantic Meridional Transect (AMT) (Aiken et al., 2000; Sal et al., 2013), and other programs provide relevant phytoplankton occurrence records, including data on species' abundance. A global synthesis of species occurrence records, including those from GBIF and OBIS has been attempted for upper trophic marine organisms, gathering 3.44 million records across nine taxa from zooplankton to sharks (Menegotto and Rangel, 2018). However, so far no effort has been undertaken to bring the various sources together for the lowest trophic marine organisms and merge them into a single harmonized database. This study aims to address this gap and to create PhytoBase, the world's largest open-ocean phytoplankton occurrence database, which may substantially reduce the global limitations associated with under-sampling.
The majority of the existing occurrence data of phytoplankton species have been collected via seawater samples of ∼5–25 mL (Lund et al., 1958; Utermöhl, 1958), followed by microscopic specimen identification. Another key source of occurrence data is the continuous plankton recorder (CPR) program, in which plankton are sampled by filtering seawater onto a silk roll (270 µm mesh size) within a recorder device that is towed behind research and commercial ships (Richardson et al., 2006). The plankton are then picked from the screens and identified by microscopy. DNA sequencing has become an alternative method to record and monitor marine phytoplankton at large scales (e.g., de Vargas et al., 2015; Sunagawa et al., 2015). However, within the recent global Tara Oceans cruise, ca. one-third of DNA sequences of plankton from seawater samples could not yet be assigned to any taxon (de Vargas et al., 2015). For the most species-rich phytoplankton group (Bacillariophyceae), 58 % of DNA sequences from seawater could be assigned to genus level in the same cruise (Malviya et al., 2016), but the majority of species have lacked reference DNA sequences needed for their identification. Additional factors have hampered the study of global phytoplankton biogeography: some surveys lack resolution in terms of the species recorded (Richardson et al., 2006; Villar et al., 2015), and abundance information in terms of cells or biomass of species is often not available in the archived records (e.g., from GBIF). Second, the taxonomic identification and chronic under-sampling of the species present in local communities via seawater samples (Cermeño et al., 2014) pose challenges that can be resolved only by trained experts or larger sampling volumes. In addition, the rapidly evolving taxonomy (e.g., Jordan, 2004) has led to varying use of nomenclature. These limitations need to be assessed and possibly overcome in a data synthesis effort.
Here, we compile 1 360 621 phytoplankton occurrence records (94.1 % resolved to the level of species; n=1704 species) and demonstrate that combining data from OBIS and GBIF increases the number of occurrence records by 52.7 % relative to the data solely obtained from OBIS. This gain increases to 65.2 % when adding occurrence data from marine surveys, including MareDat (Buitenhuis et al., 2013), AMT cruises (Sal et al., 2013), and initial Tara Oceans results (Villar et al., 2015). With respect to species abundance information, we retain cell count records whenever available from all sources, resulting in 193 763 quantitative entries. We harmonize and update the taxonomy between the sources, focusing on extant species and open-ocean records. The resulting PhytoBase dataset allows for studying global patterns in the biogeography, diversity, and composition of phytoplankton species. Using statistical SDMs, the data may serve as a starting point to examine species' niche differences across all major phytoplankton taxa and their potentially shifting distributions under climate change. The dataset can be accessed through PANGAEA, https://doi.org/10.1594/PANGAEA.904397 (Righetti et al., 2019a).
2.1 Data origin
To create PhytoBase, we compiled marine phytoplankton occurrences (i.e., presences and abundances larger than zero) from five sources, including the two largest open-access species occurrence archives: the Global Biodiversity Information Facility and the Ocean Biogeographic Information System. These two archives represent leading efforts to gather global species distribution evidence. We augmented the data with records from the Marine Ecosystem Data initiative (MareDat; Buitenhuis et al., 2013), records from a micro-phytoplankton dataset (Sal et al., 2013), and records from the global Tara Oceans cruise (Villar et al., 2015), which were not included in GBIF or OBIS at the time of data query (closing window, March 2017). While our selection of additional data was not exhaustive, it strived for the inclusion of quality-controlled large-scale phytoplankton datasets. Specifically, MareDat represents a previous global effort in gathering marine plankton data for ecological analyses (e.g., Brun et al., 2015; O'Brien et al., 2016), while Sal et al. (2013) and Villar et al. (2015) are unique in aspects of taxonomic standardization and consistency in methodology.
We retrieved occurrence records at the level “species” or below (e.g., “subspecies”, “variety” and “form”, as indicated by the taxonRank field in GBIF and OBIS downloads) for 7 phyla: Cyanobacteria, Chlorophyta (excluding macroalgae), Cryptophyta, Myzozoa, Haptophyta, Ochrophyta, and Euglenozoa. More specifically, within the Ochrophyta, we considered the classes Bacillariophyceae (diatoms), Chrysophyceae, Dictyochophyceae, Pelagophyceae, and Raphidophyceae. Within the Myzozoa, we considered the class Dinophyceae (dinoflagellates). Within the Euglenozoa, we considered the class Euglenoidea. This selection of phyla or classes strived to include all autotrophic marine phytoplankton taxa (de Vargas et al., 2015; Falkowski et al., 2004), but it is clear that some of the species may be mixotrophic, particularly for the Dinophyceae (Jeong et al., 2010). At genus level, we additionally retrieved occurrences for Prochlorococcus and Synechococcus from all sources, as the latter two genera are often highly abundant (Flombaum et al., 2013) but rarely determined to the species level. Lastly, we considered records for the functionally relevant genera Phaeocystis, Richelia, Trichodesmium, and non-specified picoeukaryotes from MareDat. For simplicity, we treat genera as species in statistics herein.
For the taxa selected, occurrence data from GBIF and OBIS were first downloaded in December 2015 and updated in February 2017. Specifically, the initial retrieval of the GBIF data occurred on 7 December 2015 (using the taxonomic backbone from https://doi.org/10.15468/39omei, last access: 14 July 2015), and the data were updated on 27 February 2017 (using an updated taxonomic backbone, accessed via http://rs.gbif.org/datasets/backbone, last access: 27 February 2017). The data from OBIS were first retrieved on 5 December 2015 using the R package robis (Provoost and Bosch, 2015) and the OBIS taxonomic backbone, accessed on 4 December 2015 via the R packages RPostgreSQL (Conway et al., 2015) and devtools (Wickham and Chang, 2015). Data were updated for the taxa selected on 6 March 2017 (using the OBIS taxonomic backbone, accessed on 6 March 2017 via the same R packages). The update in 2017 expanded the occurrences retrieved from GBIF substantially, with over 20 000 additional phytoplankton records stemming from an Australian CPR program alone (AusCPR, https://doi.org/10.1016/j.pocean.2005.09.011, accessed via https://www.gbif.org/, last access: 6 March 2017). We retained any GBIF-sourced data that were retrieved in 2015 but deleted from GBIF before March 2017 (such as CPR data, with dataset key 83986ffa-f762-11e1-a439-00145eb45e9a). Occurrence data from the Tara Oceans cruise included the Bacillariophyceae and Dinophyceae (Villar et al., 2015; their Tables W8 and W9). Occurrence data from MareDat included five phytoplankton papers (Buitenhuis et al., 2012; Leblanc et al., 2012; Luo et al., 2012; O'Brien et al., 2013; Vogt et al., 2012). Additional data processed by the Tara Oceans or Malaspina expedition (Duarte, 2015) may provide valuable context for a future synthesis and may eventually combine molecular with traditional approaches, yet here we have focused on publicly available sources up to March 2017. These sources reflect decades to centuries of efforts spent in collecting phytoplankton data, including a substantial amount of data from the CPR program (Richardson et al., 2006) and a large fraction of data from the AMT program (cruises 1 to 6) (Sal et al., 2013).
2.2 Data selection
We excluded occurrences from waters less than 200 m deep (Amante and Eakins, 2009), from enclosed seas (Baltic Sea, Black Sea, or Caspian Sea), and from seas with a surface salinity below 20, using the globally gridded (spatial ) monthly climatological data of Zweng et al. (2013). This salinity bathymetry threshold served to select data from open oceans, excluding environmentally more complex, and often more fertile, near-shore waters.
2.2.1 Data accessed through GBIF and OBIS
We included GBIF occurrence records on the basis of “human observation”, “observation”, “literature”, “living specimen”, “material sample”, “machine observation”, “observation”, and “unknown”, assuming that the latter was based on observation. With respect to OBIS data, we included data records on the basis of “O” and “D”, whereby O refers to observations and D to literature-based records. To filter out raw data of presumably inferior quality, records from OBIS and GBIF were removed: (i) if their year of collection indicated >2017 or <1800 (excluding 110 records; <0.001 % of raw data), (ii) if they had no indication on the year or month of collection (excluding 7.2 % GBIF raw data and 0.9 % OBIS raw data), or (iii) if they had geographic coordinates outside the range −180 to 180∘ for longitude and/or outside −90 to 90∘ for latitude. However, the latter criterion was fulfilled by all records, as these were standardized to −180 to 180∘ longitude (rather than 0 to 360∘ east in longitude) and −90 to 90∘ latitude (WGS84). Records with negative recording depths (0 % of GBIF and 6.6 % of OBIS raw data) were flagged and changed to positive, assuming that their original sign was mistaken.
2.2.2 Data accessed through MAREDAT
We included occurrence records at the species level for the Bacillariophyceae (Leblanc et al., 2012) and Haptophyta (O'Brien et al., 2013) and species presence records on Bacillariophyceae host cells from Luo et al. (2012). Harmonization of Haptophyta species names from MareDat (O'Brien et al., 2013) was guided by a synonymy table provided by O'Brien (personal communication, 12 June 2015) (Table A1). Harmonization of Bacillariophyceae species names in MareDat was in progress at the time of first data access (24 August 2015) and completed (Table A2). In addition, we retained all genus and species level records available for Trichodesmium, Richelia (Luo et al., 2012), Phaeocystis (Vogt et al., 2012), Synechococcus (using the data field “SynmL”), and Prochlorococcus (using the data field “PromL”) (Buitenhuis et al., 2012). We included genus level records from the latter taxa, as they represent functionally important phytoplankton groups (Le Quéré, 2005) and as information on the presence and abundance of their cells or colonial cells often only existed at genus level (Buitenhuis et al., 2012; Luo et al., 2012; Vogt et al., 2012). Across all sources, data on colonial cells could be uniquely accessed via MareDat (additional count data on trichomes of genus Trichodesmium are available from Luo et al., 2012). We also retained records of the “picoeukaryote” group, which were not determined to species or genus level (Buitenhuis et al., 2012). For all taxa, we retained records with reported abundances (i.e., cell counts) larger than zero while excluding records with zero entries or missing data entries, as our database focuses on presence only or abundance records. Given that data of the MareDat have been scrutinized previously, we flagged rather than excluded data with reported recording before the year 1800 (n=564; values 6, 10 or 11) and unrealistic day entries (n=58 340; values −9 or −1).
2.2.3 Data accessed through Villar et al. (2015)
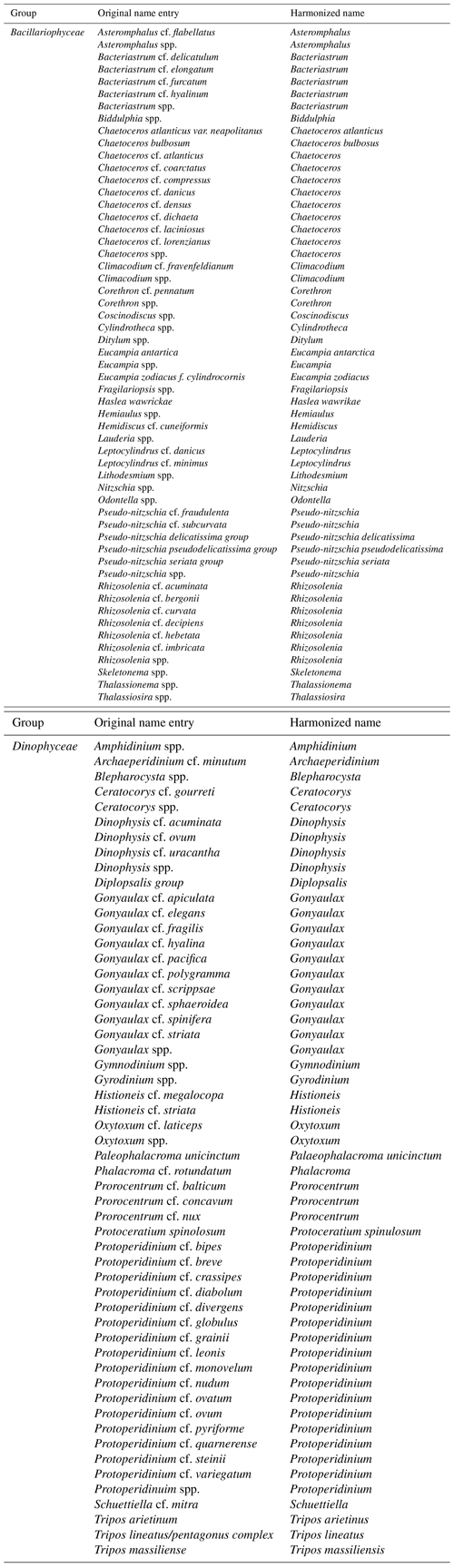
We compiled presence records of species of Bacillariophyceae and Dinophyceae from the tables W8 and W9 of Villar et al. (2015). We excluded species names containing “cf.” (e.g., Bacteriastrum cf. delicatulum), as such nomenclature is typically used to refer to closely related species of an observed species. We retained all species (n=3) that contained “group” in their names (e.g., Pseudo-nitzschia delicatissima group). Tripos lineatus/pentagonus complex was considered Tripos lineatus. The cleaning up of spelling variants of original names from Villar et al. (2015) is presented in Table A3.
2.2.4 Data accessed through Sal et al. (2013)
We considered occurrence records of the Bacillariophyceae, Dictyochophyceae, Dinophyceae, Haptophyta, and Peridinea at species level or below, using the species name in the final database. These data included 5891 records from 314 species and 543 samples. The dataset of Sal et al. (2013) represents a highly complementary source of phytoplankton occurrence records; i.e., it had no duplicated records with any of the other data sources considered. This data collection consists of in situ samples subjected to consistent methodology performed by the same taxonomist.
2.3 Concatenation of source datasets
Column names or data fields were adjusted and harmonized to establish compatibility in the dimensions of the different source datasets (Table 1). Columns match Darwin Core standard (https://dwc.tdwg.org, last access: 24 February 2020) where original data structure could be reconciled with this standard, following GBIF and OBIS, which widely rely on Darwin Core. Where critical metadata could not be assigned to Darwin Core, we use additional columns (e.g., columns ending in “gbif” present metadata from GBIF). With regard to sampling depth, GBIF raw data contained the field “depthAccuracy” (18.6 % of data with entries), while OBIS raw data contained the fields “depthprecision” (21.64 % of data with entries), “minimumDepthInMeters” (Darwin Core term; 25.7 % of data with entries), and “maximumDepthInMeters” (Darwin Core term; 24.0 % of data with entries). To enhance compatibility between GBIF and OBIS, we therefore used the column “depth”, together with “depthAccuracy”, and we integrated “depthprecision” into the latter column. To indicate the source from which records were obtained (GBIF, OBIS, MareDat, Villar or Sal) and the year of data access, we added the columns “sourceArchive” and “yearOfDataAccess”. Lastly, we added a quality flag column, termed “flag”. This column denotes records with originally negative collection depth entries (N) changed to positive (Sect. 2.2.1), unrealistic day (D) or year (Y) entries (Sect. 2.2.2), and/or records collected from sediment samples or traps (S) rather than seawater samples (Sect. 2.3.2). We concatenated the sources into a raw database, which contained 1.51 million depth-referenced occurrence records, 3300 phytoplankton species (including five genera), and 247 385 sampling events (Table 2). Sampling events are thereby (and herein) defined as unique combinations of decimal longitude, decimal latitude, depth, and time (year, month, day) in the data.
Table 1Harmonization of original column names (data fields) between data sources and final column name structure in PhytoBase.
GBIF data were downloaded in 2015 (https://www.gbif.org/, last access: 7 December 2015) and 2017 (last access: 27 February 2017) OBIS data were downloaded in 2015 (https://obis.org/, last access: 5 December 2015) and 2017 (last access: 6 March 2017). Each occurrence record in PhytoBase is uniquely identifiable by the occurrence ID: scientificName, decimalLongitude, decimalLatitude, year, month, day, and depth. a Column names following Darwin Core standard (https://dwc.tdwg.org/, last access: 24 February 2020). b We retain all original scientific name(s) and synonyms used in individual sources as additional columns with the format “scientificNameOriginal_< source>”. c The “taxonRank” field indicates the level of taxonomic resolution (species or genus) of the observation record. Records of subspecies, varieties, and forms were generally extracted from original sources but considered at the species level (using the genus and specific epithet). d Higher-order taxonomy (phylum, class) follows OBIS (taxonomic backbone; retrieved 6 March 2017), which relies on the World Register of Marine Species. e These fields indicate the organization or institution by which original records were collected. f These fields are indicators of the different research cruises or resources to which original records belonged. g The parameters “individualCount” and “observedindividualcount” had equivalent entries for records that overlapped between GBIF and OBIS and were thus merged into one column. h The parameters “datasetKey_gbif” and “resourceID_obis” are keys to access metadata of original datasets in GBIF and OBIS via API, including information on sampling methods.
2.3.1 Extant species selection and taxonomic harmonization
We strived for a selection of occurrence data of extant phytoplankton species and a taxonomic harmonization of their multiple spelling variants (merging synonyms, while clearing misspellings or unaccepted names). This procedure included three steps.
- (i)
We discarded all species (and their data) that did not have any depth-referenced record. This choice was made on the basis that these species may have been predominantly recorded via fossil materials or have been associated with large uncertainty with respect to their sampling depth.
- (ii)
We extracted all scientific names (mostly at species level, including all synonyms and spelling variants) associated with at least one depth-referenced record from the raw database (Table 2). This resulted in 3300 names, which were validated against the 150 000+ specific and infraspecific names in AlgaeBase (https://www.algaebase.org/, last access: August 2017), and matched using a relational database of current names and synonyms; orthography was made as compatible as possible with the International Code of Nomenclature (Turland et al., 2018), particularly in relation to the gender of specific epithets. This screening led to the exclusion of 459 names (and their data), which could not be traced back to any taxonomically accepted name at the time of query, and to the creation of a “synonymy table” in which each original name (including its potentially multiple synonyms and spelling errors) was matched to a corrected or accepted name.
- (iii)
We excluded species (and their data) classified as “fossil only” or “fossil” on AlgaeBase (https://www.algaebase.org/, last access: August 2017) or the World Register of Marine Species (WoRMS; http://www.marinespecies.org/, last access: August 2017). We further excluded species belonging to genera with fossil types denoted by AlgaeBase, under the condition that these species lacked habitat information on AlgaeBase, assuming that the latter species have been collected based on sedimentary or fossilized materials. Species uniquely classified as “freshwater” on AlgaeBase were discarded, as these were beyond the scope of our open-ocean database. However, we retained species classified as freshwater, which had at least 24 open-ocean (Sect. 2.2) records and thus were assumed to thrive also in marine habitats: Aulacoseira granulata, Chaetoceros wighamii, Diatoma rhombica, Dinobryon balticum, Gymnodinium wulffii, Tripos candelabrum, and Tripos euarcuatus. These simplifying steps led to a remaining set of 2032 original species names, synonyms, or spelling variants, corresponding to 1709 taxonomically harmonized species (including five genera not resolved to species level).
Table 2Summary statistics of the raw database by source.
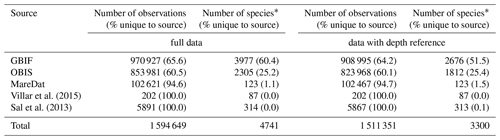
Numbers of observations (with % of observations unique to the source in parentheses) and the numbers of species (with % of species unique to the source in parentheses) presented for each data source. A total of 27 537 observation records of picoeukaryotes (not identified to species or genus level) are included among the total records and stem from MareDat (all of which contained a depth reference). ∗ Including synonyms or spelling variants.
2.3.2 Data merger and synthesis
We removed duplicate records, considering the columns “scientificName”, “decimalLongitude”, “decimalLatitude”, “year”, “month”, “day”, and “depth”. Removing duplicates meant that any relevant metadata of the duplicated (and hence removed) records were added to the metadata of the record retained, either in an existing or additional column (e.g., information on the original dataset keys to which the merged records belonged). We assigned the corrected and/or harmonized taxonomic species name to each original species name in the database on the basis of the synonymy table. We removed duplicates with respect to exact combinations of the harmonized “scientificName”, and “decimalLongitude”, “decimalLatitude”, “year”, “month”, “day”, “depth”. This resulted in the harmonized database containing 1 360 621 occurrence records (of which 95.8 % had a depth reference), 1709 species (including five genera), and 242 074 sampling events (Table 3). We retained meta-information on the dataset ID, cruise number, and further attributes when removing duplicates. In particular, we retained the original taxonomic name(s) associated with each record in separate columns of the type “scientificNameOriginal_<source>”, which allows for tracing back the harmonized name to its original name(s). Retaining original names ensures that future taxonomic changes or updated methods can be readily implemented. Aside from the presence data, the final database includes 193 777 count records of individuals or cells, spanning 1126 species. Among these, 105 242 records included a volume basis (spanning 335 species), with a predominant origin from MareDat (n=99 498) and Sal et al. (2013) (n=5744). Lastly, we flagged sedimentary records, indicated by the column “flag”. Although we probably excluded many records based on fossil materials during cleaning step (i), this does not avoid the possibility that occurrence records of extant species in the GBIF and OBIS source datasets originated partially from sediment traps or sediment core samples.
Table 3Summary statistics of the harmonized database by source.
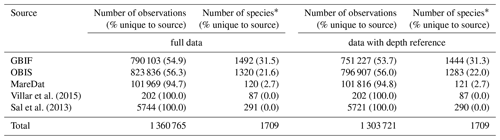
Numbers of observations (with % of observations unique to the source in parentheses) and numbers of species (with % of species unique to the source in parentheses) presented for each data source. ∗ Including 1711 species names and the genera Phaeocystis, Trichodesmium, Richelia, Prochlorococcus, and Synechococcus. A total of 27 537 observation records of picoeukaryotes (not identified to species or genus level) are included among the total records and stem from MareDat (all of which contained a depth reference).
Marine sediments can conserve phytoplankton cells that are exported to depth. We flagged phytoplankton records from OBIS and GBIF in the database associated with surface sediment traps or sediment cores (using an “S” in the flag column) by checking the metadata of each individual source dataset of GBIF (using the GBIF datasetKey) and OBIS (using the OBIS resourceID), using the function datasets in the R package rgbif (Chamberlain, 2015) and the online portal of OBIS (http://iobis.org/explore/#/dataset, last access: 24 October 2018). This check resulted in the flagging of 2.7 % of records. We did not attempt to clean or remove sediment type records in MareDat, assuming that information on sampling depth associated with records of MareDat led to thorough exclusion of sedimentary records previously. Data from Sal et al. (2013) and Villar et al. (2015) were uniquely based on seawater samples.
Data
Spatiotemporal coverage
Phytoplankton occurrence records contained in PhytoBase cover all ocean basins, latitudes, longitudes, and months (Fig. 1). However, data density is globally highly uneven (Fig. 1b, c; histograms) with 44.7 % of all records falling into the North Atlantic alone, while only 1.4 % of records originate from the South Atlantic and large parts of the South Pacific basin are devoid of records (Fig. 1a). Analyzing the data by latitude (Fig. 1b)sampling has been particularly and longitude (Fig. 1c) reveals that sampling has been particularly thin at high latitudes (>70∘ N and S) during wintertime. Occurrences cover a total of 18 863 monthly cells of 1∘ latitude × 1∘ longitude (using the World Geodetic System of 1984 as the reference coordinate system; WGS 84), which corresponds to 3.9 % of all monthly (n=12 months) 1∘ cells of the open ocean (Sect. 2.2). Without monthly distinction, records cover 6098 spatial 1∘ cells, which is a fraction of 15.5 % of all 1∘ cells of the open ocean.
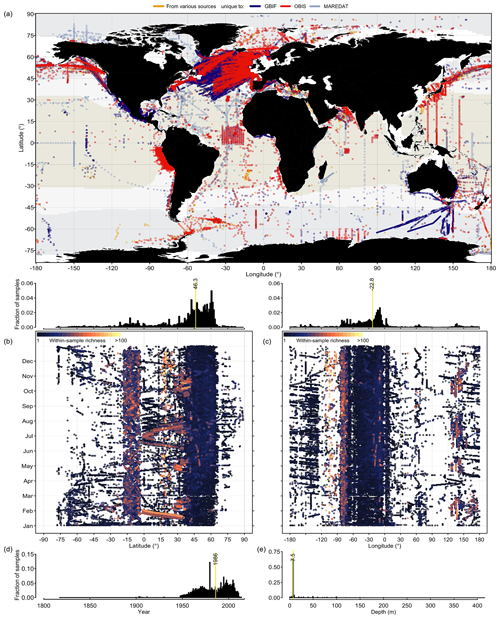
Figure 1Global distribution of phytoplankton occurrence records of PhytoBase. (a) Circles show the position of in situ occurrence records (n=1 360 765, including 1 280 103 records at the level of species), with the color indicating the source of the data. Color shading indicates the extent of tropical (T>20 ∘C; yellow), temperate (10 ∘C ≤ T≤20 ∘C; snow white), and cold (T<10 ∘C; light blue) seas, based on the annual mean sea surface temperature (Locarnini et al., 2013). (b–c) Sampling locations (dots) are plotted as a function of the month of sampling and (b) latitude or (c) longitude. Colors display the species number detected in individual samples (each sample is defined as an exact combinations of time, location, and depth in the dataset). Histograms above panels (b) and (c) show the frequency of samples by latitude (b) or longitude (c). (d–e) Histograms of sample frequency by year (d) and depth (e). Vertical yellow lines show the median.
Record quantities are not evenly distributed between major taxa, and global sampling schemes differ between these taxa (Fig. 2). CPR observations are highly condensed in the North Atlantic (and to a lesser extent south of Australia) for the Bacillariophyceae and Dinophyceae (Fig. 2a, b), but this aggregation is less clear for the Haptophyta (Fig. 2c), whose species typically have much smaller cells (often <10 µm) than the species of the former two groups. These three principal phytoplankton taxa have been surveyed well along the north–south AMT cruises, but they lack data in large areas of the South Pacific. Among the less species-rich taxonomic groups, including Cyanobacteria and Chlorophyta, global occurrence data coverage has been sparser (Fig. 2d, e). Since all of the principal phytoplankton taxa are globally abundant and widespread, the absence of records likely reflects a lack of sampling efforts rather than a lack of phytoplankton.
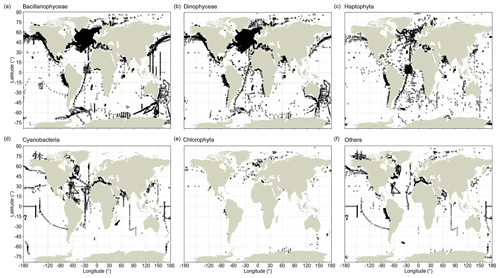
Figure 2Global distribution of phytoplankton occurrence records in PhytoBase for individual taxa. Black circles show the distribution of in situ records for the five largest phyla or classes in the database, which constitute 97.6 % of all records, (a–e) and for the remaining taxa (f). Records may overlap at any particular location.
Environmental coverage
The phytoplankton occurrences compiled cover the entire temperature range and a broad part of nitrate and mixed-layer conditions found in the global ocean (Fig. 3a, b). To visualize the environmental data coverage, Fig. 3 matches occurrence records of PhytoBase with climatological sea surface data on nitrate (Garcia et al., 2013), temperature (Locarnini et al., 2013), and mixed-layer depth (de Boyer Montégut, 2004) at monthly resolution. Records are concentrated in areas with intermediate conditions, which are relatively more frequent at the global scale (gray shade; Fig. 3a, b). Data on cell counts (7.7 % of total) show a similar coverage to the full data (Fig. 3a, b) but are much thinner (Fig. 3c, d).
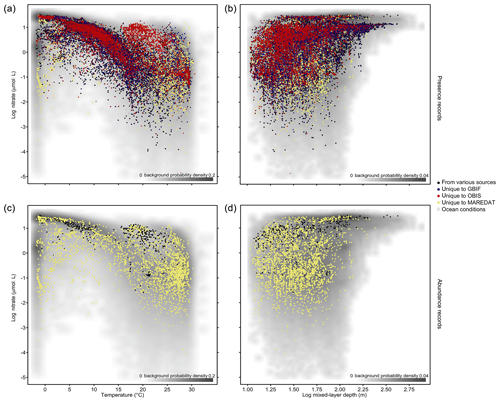
Figure 3Phytoplankton records in environmental parameter space. (a–b) Dots display in situ records (n=1 360 621) as a function of sea temperature and nitrate concentration (a) and as a function of mixed-layer depth (MLD) and nitrate concentration (b). The scale is logarithmic for MLD and nitrate. Shading indicates the frequency of environmental conditions appearing in the open ocean at the surface, with a darker gray shade indicating higher frequency of occurrence (bivariate Gaussian kernel density estimate). The colors of the dots denote the source of data, indicating complementarity or overlap of the environmental gradients sampled between sources. Panels (c–d) show the subset of records that contain information on species' cell counts per liter (n=105 242) stemming largely from MareDat.
Taxonomic coverage
We assessed what fraction of the known marine phytoplankton species (Falkowski et al., 2004; Jordan, 2004; de Vargas et al., 2015) is represented in PhytoBase. The records include all major marine taxa of phytoplankton known (n=16 classes), including Bacillariophyceae, Dinophyceae, and Haptophyta. Records span roughly half of the known marine species of the Haptophyta (Jordan, 2004) and a similar fraction of the known marine species of Bacillariophyceae and Dinophyceae (Table 4). By contrast, species of the less species-rich taxa tend to be more strongly underrepresented and account for a relatively small fraction (<3 %) of all species in PhytoBase.
Record quantities are unevenly distributed between individual species (Fig. 4). Half of the species contain at least 30 presence records, but multiple species contribute one or two records (Fig. 4a). The species with fewer than 30 records account for as little as 0.54 % of all species records in PhytoBase. Similarly, half of all genera contain at least 110 records each, while genera with fewer than 110 records each contribute as little as 8.2 % to the total of records. A similar data distribution applies to the subset of species (n=330) for which cell count records (with a volume basis) are available (Fig. 4b). Half of these species contribute at least 16 records, and among the genera containing cell counts, half contribute at least 76 records.
Completeness of species richness inventories at large spatial scales
We analyzed the ocean inventory of phytoplankton species richness in the database for three different regimes of ocean temperature by means of species accumulation curves (SACs) (Thompson and Withers, 2003) (Fig. 5). These curves present the cumulative species richness detected as a function of sampling effort (or survey area) and are expected to increase asymptotically before they saturate above a certain threshold of sampling effort (i.e., when the system has been exhaustively sampled). Using the number of sampling events (i.e., unique combinations of time, depth, location in our database) as a surrogate for sampling effort (x axis), we find that the richness detected (y axis) and the completeness of species richness detection (degree of saturation) differ notably between regimes. In the southern temperate (Fig. 5e) and cold seas (Fig. 5f), species richness has been incompletely sampled with respect to all taxa (black lines) or key taxa (colored lines). By contrast, SACs in the Northern Hemisphere start to saturate at ∼40 000 samples, suggesting that the sampling has recorded a majority of the species. Specifically, SACs suggest that species richness will saturate at around ∼1500 species in the tropical regime (>20 ∘C), at ∼1100 species in northern midlatitudes (≥10 ∘C, ≤20 ∘C), and at ∼600 species in the cold northern seas (<10 ∘C). This corresponds to 93 %, 64 %, and 35 % of all ∼1700 species collected in PhytoBase, respectively. However, these estimates only represent the fraction of species detectable via light microscopy and other methods underlying our database, preferentially omitting very rare or small species (Cermeño et al., 2014; Ser-Giacomi et al., 2018; Sogin et al., 2006). Thus, the richness will likely increase (at low rates) with additional sampling effort. Theoretical models have suggested that communities with many rare species lead to SACs with “low shoulders”, meaning that SACs have a long upward slope to the asymptote (Thompson and Withers, 2003), consistent with our SACs (Fig. 5).
Table 4Statistics on the number of records and species contained in the database for key taxa.
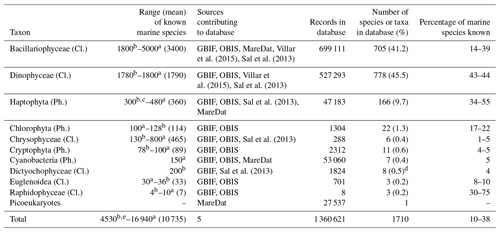
Cl. is an abbreviation for class, and Ph. is an abbreviation for phylum.
The table summarizes the occurrence records for the 10 major taxa in
PhytoBase and describes to what
degree the species in each taxon represent
the total number of marine species known (for which exact numbers are still
debated; we therefore provide upper and lower bounds
and mean values in
parentheses).
a Falkowski et al. (2004); this estimate includes both
coastal and open-ocean taxa, while PhytoBase focuses primarily on data from
the open
ocean.
b de Vargas et al. (2015).
c Jordan et al. (2004).
d Including one species of the sister class Pelagophyceae.
e The estimate by de Vargas et al. (2015) excluded
prokaryotes.
A number of 150 prokaryotes (Falkowski et al., 2004) were added
to obtain the mean.

Figure 4Distribution of occurrence records between species or genera. Histograms show the frequency of species (black) and genera (yellow) with a certain amount of (a) presence or (b) abundance records, separately. Vertical lines (black, yellow) indicate the median value. The x axes are logarithmic to the base 10.
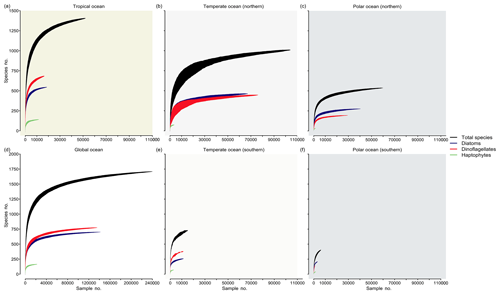
Figure 5Accumulation of species richness as a function of sampling effort by region. Curves show the cumulative species richness as a function of samples (i.e., unique combinations of space, time, and depth in the database drawn at random) drawn at random from the database, using 100 runs (shadings around the curves indicate ± 1 SD). Shown are species accumulation curves for all species (black) and three major taxa (colors) for (a) the tropics, defined as regions with a sea surface temperature (T) >20 ∘C; (b) temperate seas (10 ∘C ≤ T≤20 ∘C) of the Northern Hemisphere; (c) cold seas (T<10 ∘C) of the Northern Hemisphere; (d) global ocean; (e) temperate seas (10 ∘C ≤ T≤20 ∘C) of the Southern Hemisphere; and (f) cold seas (T<10 ∘C) of the Southern Hemisphere. Background colors refer to Fig. 1A.
Species richness documented within 1∘ cells
To explore how completely species richness has been sampled at much smaller spatial scales, we binned data at 1∘ × 1∘ resolution and analyzed the number of species in the pooled data per cell as a function of sampling effort. Hotspots in directly observed phytoplankton richness at the 1∘ cell level emerge in near-shore waters of Peru, around California, southeast of Australia, in the North Atlantic, along AMT cruises, and along research transects south of Japan (Fig. 6a). The species richness detected per 1∘ cell is positively correlated with sampling effort, using the number of samples collected per cell as a surrogate of sampling effort (Spearman's ρ=0.47, P<0.001). In particular, the richness of Bacillariophyceae (ρ=0.88, P<0.001) and Dinophyceae (ρ=0.92, P<0.001) is positively correlated with effort, while this is less the case for the Haptophyta (ρ=0.27; P<0.001). Analyzing species richness as a function of “sampling events” for different thermal regimes separately reveals that tropical areas (yellow dots; Fig. 6b–e) yield higher cumulative per cell richness at moderate to high sampling effort (>50 samples) than temperate (gray dots) and polar areas (blue dots). Although data are thin and scattered, species richness in cold areas tends to saturate at ∼70 species per cell (Fig. 6b; blue dots) at an effort of ∼500 samples collected per cell. In contrast, species richness of the tropical areas tends to reach ∼290 species per cell at the same effort (∼500 samples). This suggests that tropical phytoplankton richness at the cell level is about 4 times higher than that of cold northern areas, but richness may further increase with additional sampling effort. Analyzing the data of the major taxa separately suggests that roughly 200 species of Bacillariophyceae and Dinophyceae can be collected per cell at high sampling effort (∼500 samples), yet data are sparse for Haptophyta, which broadly lack 1∘ cells with more than 100 samples collected (Fig. 6e).
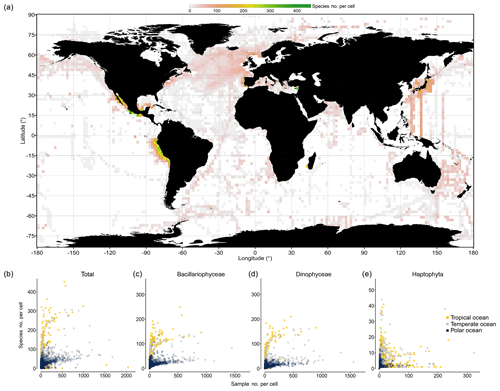
Figure 6Species richness observed within 1∘ cells. (a) Global map visualizing the species richness detected within each 1∘ latitude × 1∘ longitude cell of the ocean. (The means of four 1∘ cells are depicted at 2∘ resolution). (b–e) The number of species detected within each 1∘ cell is plotted as a function of sampling effort (i.e., number of sampling events, defined as unique combinations of position, time, and depth in the database), with colors indicating data originating from different regions: tropical (T>20 ∘C; yellow), temperate (10 ∘C ≤ T≤20 ∘C; snow white), and polar 1∘ cells (T<10 ∘C; light blue), as defined by the annual mean temperature at sea surface (Locarnini et al., 2013; see shading of map in Fig. 1). The richness–effort relationship is shown for all taxa (b) and major taxa separately (c–e).
The analysis of species richness detected per 1∘ cell suggests that approximately one-third to one-fifth of all species inventoried in the tropical or polar regime (see Fig. 5) through our database can be detected within a single 1∘ cell of these regimes at high sampling effort (∼500 samples) (Fig. 6b). This result is in coarse agreement with the result obtained at the large spatial scale (Fig. 5a–c), where the richness detected in the tropical regime was close to 3 times that of the (northern) cold regime.
Comparative spatial and taxonomic analysis of source datasets
We used the sources obtained from within the GBIF archive as an exemplary case for a more detailed examination of original source dataset coverage, as GBIF provides relatively detailed information on its sources via dataset keys. CPR is the single largest source dataset obtained from GBIF, which covers the North Atlantic and North Pacific (Fig. 7a–d; brown dots) and parts of the ocean south of Australia (Fig. 7a–d; blue dots). CPR records obtained via GBIF contribute 33.9 % to all records in PhytoBase. CPR data show relatively low species numbers captured on average per “sample” (Fig. 7i), with samples being defined as exact combinations of geographic position, depth, and time in the data records. This may be owing to the continuous collection of species or incomplete reporting of taxa. The mesh size of the silk employed in CPR of 270 µm under-samples small phytoplankton species (<10 µm). However, small species nevertheless get regularly captured in CPR, as they get attached to the screens (Richardson et al., 2006). Within the 16 largest source datasets obtained via GBIF, the average number of species collected per sample is below 4 for the CPR program and increases to more than 50 for other datasets (Fig. 7i). These 16 test datasets (excluding datasets containing sedimentary records) highlight that the taxonomic resolution strongly differs between samples of individual cruises or survey programs. By latitude, different surveys or cruises thus contribute to PhytoBase to a varying degree (Fig. 7e–h). Systematic differences in the species detected per sample and the varying contribution of sources to the database along latitude (Fig. 7e–h) are important considerations when, for example, analyzing species richness directly.
Analyzing the 16 largest source datasets from GBIF in environmental parameter space (Fig. 8) reveals the association of individual datasets with subdomains of the global temperatures, nitrate levels or mixed-layer depths. GBIF datasets collected in the tropics and subtropics (mean temperature of sampling ≥20 ∘C; Fig. 8a) tend to be associated with higher taxonomic detail (∼25 species detected per sample on average; Fig. 7i), compared to datasets collected in colder areas. However, this likely also reflects an overall higher number of species occurring in tropical areas (Fig. 5a) than extratropical ones.
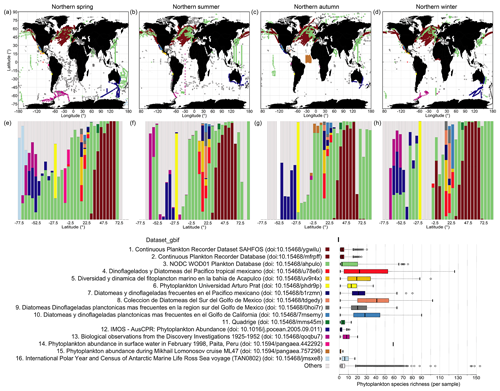
Figure 7Spatial extent of the 16 largest datasets from GBIF and average richness per sample. (a–d) Maps display the spatial distribution of the 16 largest contributing datasets to the GBIF-sourced data in PhytoBase, showing each season separately. The datasets presented comprise 54.3 % of all records and 93.5 % of GBIF-sourced records. GBIF data are shown as an exemplary case, as GBIF contributes a variety of source datasets referenced by dataset keys (datasetKey_gbif). Panels (e–h) show the relative contribution of datasets to the occurrence data per 5∘ latitude. Colored sub-bars represent specific datasets (see i), and their widths show the amount of data contributed. Panels (e–h) present the data shown in (a–d). (i) Analysis of within-sample species richness. Boxes show the mean species richness (thick vertical lines) detected per sample of specific datasets and the first and third quartiles around the mean (boxes). Whiskers denote 2.5 times the interquartile range. Note that the same analysis may be performed for OBIS data using the field “resourceID_obis”.
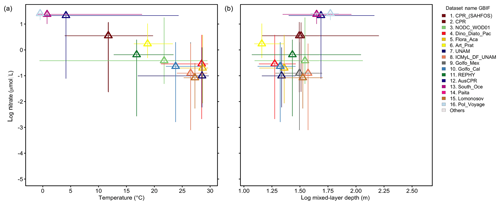
Figure 8Environmental range of the 16 largest datasets from GBIF. (a–b) The range of 16 datasets contained within GBIF-sourced data are represented by thin lines in parameter space: (a) temperature vs. logarithmic nitrate concentration in the surface ocean and (b) logarithmic mixed-layer depth vs. logarithmic nitrate (using climatological environmental data from Garcia et al., 2013; Locarnini et al., 2013; de Boyer Montégut, 2004; matched with records at monthly climatological 1∘ resolution). Lines span the minimum to maximum environmental conditions associated with the records of each dataset separately. Triangles display the mean environmental condition of the records per dataset.
Sensitivity of data to taxonomic harmonization and coordinate rounding
Compared to OBIS, GBIF contributed roughly 14 % additional records to the raw database (Table 2), yet this relative contribution changed after the harmonization step of species names. GBIF and OBIS finally contributed 790 103 and 823 836 records, respectively, to the harmonized PhytoBase. Hence, the exclusion of nonmarine, fossil, or doubtful species and the taxonomic harmonization step were more stringent for GBIF-sourced records than OBIS-sourced records.
We tested to what degree the number of unique records in the harmonized database changed when decimal positions in the coordinates of each of the five data sources were rounded prior to their merger. We find that the total number of unique records in PhytoBase declines continuously from 1.36 million to 1.07 million, when rounding the coordinates of records in the data to the sixth, fifth, fourth, third, and second decimal place. This result may be explained by the fact that large parts of the data come from CPR. The records of CPR start to be binned into coarser sampling units when rounding their decimal positions. The harmonized database (without coordinate rounding) contained 65.2 % additional records compared to its largest contributing source. This gain was similarly high in the non-harmonized raw database and increased to ca. 73 % when rounding coordinates to varying decimals. This shows that different sources contributed highly complementary records to PhytoBase, regardless of a coordinate rounding to varying decimals.
4.1 Data coverage, uncertainties, and recommendations
Spatiotemporal data on species occurrence are an essential basis to assess and forecast species' distributions and to understand the drivers behind these patterns. Following recent calls to gather species occurrences into global databases (Edwards, 2000; Meyer et al., 2015), we merged occurrence data of marine phytoplankton from three data sources and from the two largest open-access biological data archives into PhytoBase. This new database contains 1 360 621 records (1 280 103 records at the level of species), describing 1716 species of seven phyla. Our effort addresses a gap in marine species occurrence data, as previous studies of marine taxa (Tittensor et al., 2010; Chaudhary et al., 2016; Menegotto and Rangel, 2018) had no easy access to data sufficiently complete for global analyses of phytoplankton. The synthesis and harmonization of GBIF data with OBIS and other sources results in a substantial gain of phytoplankton occurrence data (>60 % additional data) relative to phytoplankton data residing in either of the two archives. The harmonization of different archives, which collect global species distribution evidence, therefore substantially expanded the empirical basis of phytoplankton records.
PhytoBase presents, to our knowledge, the largest current global database of marine phytoplankton species occurrences. However, two main limitations remain: first, the global data density is highly uneven spatially, and gaps persist across large swaths of the ocean, e.g., in the South Pacific Ocean and the central Indian Ocean. Second, sampling priorities with respect to taxonomic groups, size classes or species resolution differ widely between research cruises and programs. While small or fragile species may escape detection by the CPR program (Richardson et al., 2006), the resolution of traditional samples is influenced by sampling volume and taxonomic expertise (Cermeño et al., 2014). Our results show that the average number of species detected per sample varies from 3 to above 50 between different cruises or programs. A global spatial bias in collection density of marine species has similarly been found for heterotrophic taxa (Menegotto and Rangel, 2018), but sampling biases and divergent sampling protocols may be even more common for phytoplankton.
Owing to these limitations, we recommend that direct analyses of the database be undertaken and interpreted with caution. For example, our data analysis has shown that direct species richness estimates are sensitive to the number of sampling events. In addition, many species have low occurrence numbers in the database, making any inference about their ecological niche or geographic distribution very uncertain. Thus, without careful screening and checking of the data (via, e.g., datasetKeys for GBIF records and resourceIDs for OBIS records), the characterization of biogeographies at the species level might be highly biased.
Statistical techniques such as rarefaction (Rodríguez-Ramos et al., 2015), randomized resampling (Chaudhary et al., 2017), analysis of sampling gaps (Woolley et al., 2016; Menegotto and Rangel, 2018), and species distribution modeling (Zimmermann and Guisan, 2000) may be implemented to overcome these limitations. The latter statistical technique may be particularly promising, as species distribution models can be set up to account for variation in presence data sampling (Phillips et al., 2009) and data scarceness (Breiner et al., 2015). Based on observed associations between species' occurrences and environmental factors (Guisan and Thuiller, 2005), these models estimate the species' ecological niche, which is projected into geographic space, assuming that the species' niche and its geographic habitat are directly interrelated (Colwell and Rangel, 2009). Another advantage of species distribution models is that they can circumvent geographic sampling gaps through a spatial projection of the niche, as long as environmental conditions relevant to describe the niche of the species have been sufficiently well sampled and the species fills its ecological niche. This is the approach used by Righetti et al. (2019b), building on a large fraction of the PhytoBase (77.6 % of the records, falling into the monthly climatological mixed-layer; de Boyer Montégut, 2004), to analyze global richness patterns of phytoplankton.
The detection of rare species and their integration into PhytoBase may become possible via molecular methods (Bork et al., 2015; Sogin et al., 2006). DNA sequencing has become an alternative approach to characterize phytoplankton biogeography (de Vargas et al., 2015). These data have two advantages over traditional taxonomic data: first, the sensitivity of metagenomic methods to detect rare taxa is much higher relative to traditional data. Second, metagenomic data have been collected in a methodologically consistent way in recent global surveys, such as the Tara Oceans cruise (de Vargas et al., 2015). However, there are also drawbacks associated with DNA-based methods. A large disadvantage of current metagenomic data is the lack of catalogued reference gene sequences for most species. As a result, the majority of the metagenomic sequences can only be determined to the level of genus (Malviya et al., 2016). However, we expect that an integration of detailed genetic data with traditional sampling data may soon become possible, allowing us to combine several methodological or taxonomic dimensions in databases. At any point in the future, changing taxonomic nomenclature may be readily implemented, as we retained the original names and synonyms from raw data sources together with the harmonized name for each record in PhytoBase.
4.2 Data use
Our data compilation and synthesis product PhytoBase has been designed to primarily support the analysis of the distribution, diversity, and abundance of phytoplankton species and related biotic or abiotic drivers in macroecological studies. However, PhytoBase is far from limited to this set of applications and may include the analysis of ecological niche differences between species or clades, linkages between species' ecological niches and phylogenetic or functional relatedness, current or future spatial projections of species' niches and composition, tests on whether presence–absence patterns of multiple species can predict community trait indices, or joint analyses of species' distribution and trait data to project trait biogeographies. The database may also be used to validate the increasingly complex marine ecosystem models included in regional to global climate models.
The accuracy of data analyses may be limited by sampling biases underlying PhytoBase, including the spatiotemporal variation in sampling efforts and varying taxonomic detail between data sources. The latter limitation might be alleviated by considering different methodologies associated with varying cruises or collecting organizations in analyses. Where possible, we thus retained the information on the original dataset ID or dataset key along with each occurrence record in the database. Moreover, statistical analysis tools may be used to address spatiotemporal variation in global sampling efforts. Given the critical under-sampling of the Southern Hemisphere, data from areas such as the South Pacific will likely lead to new species discoveries and may greatly improve the global observational basis of phytoplankton occurrences in the future. Data inclusion from recent cruises, which are still under evaluation, appears to be a natural next step. These data may come from the Malaspina expedition (Duarte, 2015), Tara Oceans (Bork et al., 2015) and Southern Ocean transects (Balch et al., 2016).
PhytoBase is publicly available through PANGAEA, https://doi.org/10.1594/PANGAEA.904397 (Righetti et al., 2019a). Associated R scripts and the synonymy table used to harmonize species' names are available through https://gitlab.ethz.ch/phytobase/supplementary (last access: 20 April 2020).
In PhytoBase, we compiled more than 1.36 million marine phytoplankton records that span 1704 species, including the key taxa Bacillariophyceae, Dinophyceae, Haptophyta, Cyanobacteria, and others. The database addresses photosynthetic microbial organisms, which play crucial roles in global biogeochemical cycles and marine ecology. We have provided an analysis of the current status of marine phytoplankton occurrence records accessible through public archives, their spatial and methodological limitations, and the completeness of species richness information for different ocean regions. PhytoBase may stimulate studies into the biogeography, diversity, and composition of phytoplankton and serve to calibrate ecological or mechanistic models. We recommend accounting carefully for data structure and metadata, depending on the purpose of analysis.
Table A1Harmonization of 113 taxon names contained in the data field “Species/lowest classification” of the MareDat dataset of O'Brien et al. (2013). Only the 113 names that changed during harmonization are shown out of a total of 197 names. Spaces in original taxon names are indicated by “_”.

Table A2Harmonization of 156 taxon names contained in the data-field ”Corrected name entry” of the MareDat dataset of Leblanc et al. (2012). Only the 156 names that changed during harmonization are shown out of a total of 248 names.

MV, NG, NEZ, and DR conceived the study. MDG performed the taxonomic expert screening. MV compiled substantial parts of the MareDat database. DR compiled the data, developed the code, and led the writing, with inputs by all authors.
The authors declare that they have no conflict of interest.
We thank all biologists, taxonomists, and cruise organizers for their essential efforts in collecting and sharing occurrence data of marine phytoplankton synthesized in this database. We thank Marta Estrada for her expertise on taxonomic nomenclature. We thank Markus Döring and Pieter Provoost for their support with retrieval of phytoplankton occurrence data from GBIF and OBIS.
This research has been supported by ETH Zürich (grant no. ETH-52 13-2).
This paper was edited by Francois Schmitt and reviewed by Griet Neukermans and two anonymous referees.
Aiken, J., Rees, N., Hooker, S., Holligan, P., Bale, A., Robins, D., Moore, G., Harris, R., and Pilgrim, D.: The Atlantic Meridional Transect: overview and synthesis of data, Prog. Oceanogr., 45, 257–312, https://doi.org/10.1016/S0079-6611(00)00005-7, 2000.
Amante, C. and Eakins, B. W.: ETOPO1 1 Arc-Minute Global Relief Model: Procedures, Data Sources and Analysis, NOAA Tech. Memo. NESDIS NGDC-24, Natl. Geophys. Data Center, NOAA, 2009, https://doi.org/10.7289/V5C8276M, 2009.
Balch, W. M., Bates, N. R., Lam, P. J., Twining, B. S., Rosengard, S. Z., Bowler, B. C., Drapeau, D. T., Garley, R., Lubelczyk, L. C., Mitchell, C., and Rauschenberg, S.: Factors regulating the Great Calcite Belt in the Southern Ocean and its biogeochemical significance, Global Biogeochem. Cy., 30, 1124–1144, https://doi.org/10.1002/2016GB005414, 2016.
Bork, P., Bowler, C., de Vargas, C., Gorsky, G., Karsenti, E., and Wincker, P.: Tara Oceans studies plankton at planetary scale, Science, 348, 873–873, https://doi.org/10.1126/science.aac5605, 2015.
Breiner, F. T., Guisan, A., Bergamini, A., and Nobis, M. P.: Overcoming limitations of modelling rare species by using ensembles of small models, Methods Ecol. Evol., 6, 1210–1218, https://doi.org/10.1111/2041-210X.12403, 2015.
Brun, P., Vogt, M., Payne, M. R., Gruber, N., O'Brien, C. J., Buitenhuis, E. T., Le Quéré, C., Leblanc, K., and Luo, Y.-W.: Ecological niches of open ocean phytoplankton taxa, Limnol. Oceanogr., 60, 1020–1038, https://doi.org/10.1002/lno.10074, 2015.
Buitenhuis, E. T., Li, W. K. W., Vaulot, D., Lomas, M. W., Landry, M. R., Partensky, F., Karl, D. M., Ulloa, O., Campbell, L., Jacquet, S., Lantoine, F., Chavez, F., Macias, D., Gosselin, M., and McManus, G. B.: Picophytoplankton biomass distribution in the global ocean, Earth Syst. Sci. Data, 4, 37–46, https://doi.org/10.5194/essd-4-37-2012, 2012.
Buitenhuis, E. T., Vogt, M., Moriarty, R., Bednaršek, N., Doney, S. C., Leblanc, K., Le Quéré, C., Luo, Y.-W., O'Brien, C., O'Brien, T., Peloquin, J., Schiebel, R., and Swan, C.: MAREDAT: towards a world atlas of MARine Ecosystem DATa, Earth Syst. Sci. Data, 5, 227–239, https://doi.org/10.5194/essd-5-227-2013, 2013.
Cermeño, P., Teixeira, I. G., Branco, M., Figueiras, F. G., and Marañón, E.: Sampling the limits of species richness in marine phytoplankton communities, J. Plankton Res., 36, 1135–1139, https://doi.org/10.1093/plankt/fbu033, 2014.
Chamberlain, S.: rgbif: Interface to the Global Biodiversity Information Facility API, R package version 0.9.7, 2015.
Chaudhary, C., Saeedi, H., and Costello, M. J.: Bimodality of latitudinal gradients in marine species richness, Trends Ecol. Evol., 31, 670–676, https://doi.org/10.1016/j.tree.2016.06.001, 2016.
Chaudhary, C., Saeedi, H., and Costello, M. J.: Marine species richness is bimodal with latitude: a reply to fernandez and marques, Trends Ecol. Evol., 32, 234–237, https://doi.org/10.1016/j.tree.2017.02.007, 2017.
Colwell, R. K. and Rangel, T. F.: Hutchinson's duality: The once and future niche, P. Natl. Acad. Sci. USA, 106, 19651–19658, https://doi.org/10.1073/pnas.0901650106, 2009.
Conway, J., Eddelbuettel, D., Nishiyama, T., Prayaga, S. K., and Tiffin, N.: RPostgreSQL: R interface to the PostgreSQL database system. R package version 0.4, 2015.
de Boyer Montégut, C.: Mixed layer depth over the global ocean: An examination of profile data and a profile-based climatology, J. Geophys. Res., 109, C12003, https://doi.org/10.1029/2004JC002378, 2004.
de Vargas, C., Audic, S., Henry, N., Decelle, J., Mahe, F., Logares, R., Lara, E., Berney, C., Le Bescot, N., Probert, I., Carmichael, M., Poulain, J., Romac, S., Colin, S., Aury, J.-M., Bittner, L., Chaffron, S., Dunthorn, M., Engelen, S., Flegontova, O., Guidi, L., Horak, A., Jaillon, O., Lima-Mendez, G., Luke, J., Malviya, S., Morard, R., Mulot, M., Scalco, E., Siano, R., Vincent, F., Zingone, A., Dimier, C., Picheral, M., Searson, S., Kandels-Lewis, S., Acinas, S. G., Bork, P., Bowler, C., Gorsky, G., Grimsley, N., Hingamp, P., Iudicone, D., Not, F., Ogata, H., Pesant, S., Raes, J., Sieracki, M. E., Speich, S., Stemmann, L., Sunagawa, S., Weissenbach, J., Wincker, P., Karsenti, E., Boss, E., Follows, M., Karp-Boss, L., Krzic, U., Reynaud, E. G., Sardet, C., Sullivan, M. B., and Velayoudon, D.: Eukaryotic plankton diversity in the sunlit ocean, Science, 348, 1261605–1261605, https://doi.org/10.1126/science.1261605, 2015.
Duarte, C. M.: Seafaring in the 21St Century: The Malaspina 2010 Circumnavigation Expedition, Limnol. Oceanogr. Bull., 24, 11–14, https://doi.org/10.1002/lob.10008, 2015.
Edwards, J. L.: Interoperability of biodiversity databases: biodiversity information on every desktop, Science, 289, 2312–2314, https://doi.org/10.1126/science.289.5488.2312, 2000.
Endo, H., Ogata, H., and Suzuki, K.: Contrasting biogeography and diversity patterns between diatoms and haptophytes in the central Pacific Ocean, Sci. Rep., 8, 10916, https://doi.org/10.1038/s41598-018-29039-9, 2018.
Falkowski, P. G., Katz M. E., Knoll, A. H., Quigg, A., Raven, J. A., Schofield, O., and Taylor, F. J. R.: The evolution of modern eukaryotic phytoplankton, Science, 305, 354–360, https://doi.org/10.1126/science.1095964, 2004.
Field, C. B., Behrenfeld, M. J., Tanderson, J. T., and Falkowski, P.: Primary production of the biosphere: Integrating terrestrial and oceanic components, Science, 281, 237–240, https://doi.org/10.1126/science.281.5374.237, 1998.
Flombaum, P., Gallegos, J. L., Gordillo, R. A., Rincon, J., Zabala, L. L., Jiao, N., Karl, D. M., Li, W. K. W., Lomas, M. W., Veneziano, D., Vera, C. S., Vrugt, J. A., and Martiny, A. C.: Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus, P. Natl. Acad. Sci. USA, 110, 9824–9829, https://doi.org/10.1073/pnas.1307701110, 2013.
Garcia, H. E., Locarnini, R. A., Boyer, T. P., Antonov, J. I., Baranova, O. K., Zweng, M. M., Reagan, J. R., and Johnson, D. R.: World Ocean Atlas 2013, Vol. 4 Dissolved Inorg. Nutr. (phosphate, nitrate, silicate), edited by: Levitus, S. and Mishonov, A., 25, 2013.
Guisan, A. and Thuiller, W.: Predicting species distribution: Offering more than simple habitat models, Ecol. Lett., 8, 993–1009, https://doi.org/10.1111/j.1461-0248.2005.00792.x, 2005.
Guisan, A. and Zimmermann, N. E.: Predictive habitat distribution models in ecology, Ecol. Modell., 135, 147–186, https://doi.org/10.1016/S0304-3800(00)00354-9, 2000.
Honjo, S. and Okada, H.: Community structure of coccolithophores in the photic layer of the mid-pacific, Micropaleontology, 20, 209–230, https://doi.org/10.2307/1485061, 1974.
Iglesias-Rodríguez, M. D., Brown, C. W., Doney, S. C., Kleypas, J., Kolber, D., Kolber, Z., Hayes, P. K., and Falkowski, P. G.: Representing key phytoplankton functional groups in ocean carbon cycle models: Coccolithophorids, Global Biogeochem. Cy., 16, 471–4720, https://doi.org/10.1029/2001GB001454, 2002.
Jeong, H. J., Yoo, Y. Du, Kim, J. S., Seong, K. A., Kang, N. S., and Kim, T. H.: Growth, feeding and ecological roles of the mixotrophic and heterotrophic dinoflagellates in marine planktonic food webs, Ocean Sci. J., 45, 65–91, https://doi.org/10.1007/s12601-010-0007-2, 2010.
Jones, M. C. and Cheung, W. W. L.: Multi-model ensemble projections of climate change effects on global marine biodiversity, ICES J. Mar. Sci., 72, 741–752, https://doi.org/10.1093/icesjms/fsu172, 2015.
Jordan, R. W.: A revised classification scheme for living haptophytes, Micropaleontology, 50, 55–79, https://doi.org/10.2113/50.Suppl_1.55, 2004.
Leblanc, K., Arístegui, J., Armand, L., Assmy, P., Beker, B., Bode, A., Breton, E., Cornet, V., Gibson, J., Gosselin, M.-P., Kopczynska, E., Marshall, H., Peloquin, J., Piontkovski, S., Poulton, A. J., Quéguiner, B., Schiebel, R., Shipe, R., Stefels, J., van Leeuwe, M. A., Varela, M., Widdicombe, C., and Yallop, M.: A global diatom database – abundance, biovolume and biomass in the world ocean, Earth Syst. Sci. Data, 4, 149–165, https://doi.org/10.5194/essd-4-149-2012, 2012.
Le Quéré, C.: Ecosystem dynamics based on plankton functional types for global ocean biogeochemistry models, Global Change Biol., 11, 2016–2040, https://doi.org/10.1111/j.1365-2486.2005.1004.x, 2005.
Locarnini, R. A., Mishonov, A. V., Antonov, J. I., Boyer, T. P., Garcia, H. E., Baranova, O. K., Zweng, M. M., Paver, C. R., Reagan, J. R., Johnson, D. R., Hamilton, M., and Seidov, D.: World Ocean Atlas 2013, Vol. 1 Temp, edited by: Levitus, S. and Mishonov, A., NOAA Atlas NESDIS 73, 40, 2013.
Lund, J. W. G., Kipling, C., and Le Cren, E. D.: The inverted microscope method of estimating algal numbers and the statistical basis of estimations by counting, Hydrobiol., 11, 143–170, https://doi.org/10.1007/BF00007865, 1958.
Luo, Y.-W., Doney, S. C., Anderson, L. A., Benavides, M., Berman-Frank, I., Bode, A., Bonnet, S., Boström, K. H., Böttjer, D., Capone, D. G., Carpenter, E. J., Chen, Y. L., Church, M. J., Dore, J. E., Falcón, L. I., Fernández, A., Foster, R. A., Furuya, K., Gómez, F., Gundersen, K., Hynes, A. M., Karl, D. M., Kitajima, S., Langlois, R. J., LaRoche, J., Letelier, R. M., Marañón, E., McGillicuddy Jr., D. J., Moisander, P. H., Moore, C. M., Mouriño-Carballido, B., Mulholland, M. R., Needoba, J. A., Orcutt, K. M., Poulton, A. J., Rahav, E., Raimbault, P., Rees, A. P., Riemann, L., Shiozaki, T., SubramanTiam, A., Tyrrell, T., Turk-Kubo, K. A., Varela, M., Villareal, T. A., Webb, E. A., White, A. E., Wu, J., and Zehr, J. P.: Database of diazotrophs in global ocean: abundance, biomass and nitrogen fixation rates, Earth Syst. Sci. Data, 4, 47–73, https://doi.org/10.5194/essd-4-47-2012, 2012.
Malviya, S., Scalco, E., Audic, S., Vincent, F., Veluchamy, A., Poulain, J., Wincker, P., Iudicone, D., de Vargas, C., Bittner, L., Zingone, A., and Bowler, C.: Insights into global diatom distribution and diversity in the world's ocean, P. Natl. Acad. Sci. USA, 113, E1516–E1525, https://doi.org/10.1073/pnas.1509523113, 2016.
Mawji, E., Schlitzer, R., Dodas, E. M., and GEOTRACES-group: The GEOTRACES intermediate data product 2014, Mar. Chem., 177, 1–8, https://doi.org/10.1016/j.marchem.2015.04.005, 2015.
McQuatters-Gollop, A., Edwards, M., Helaouët, P., Johns, D. G., Owens, N. J. P., Raitsos, D. E., Schroeder, D., Skinner, J., and Stern, R. F.: The Continuous Plankton Recorder survey: How can long-term phytoplankton datasets contribute to the assessment of Good Environmental Status?, Estuar. Coast. Shelf Sci., 162, 88–97, https://doi.org/10.1016/j.ecss.2015.05.010, 2015.
Menegotto, A. and Rangel, T. F.: Mapping knowledge gaps in marine diversity reveals a latitudinal gradient of missing species richness, Nat. Commun., 9, 1–6, https://doi.org/10.1038/s41467-018-07217-7, 2018.
Meyer, C., Kreft, H., Guralnick, R., and Jetz, W.: Global priorities for an effective information basis of biodiversity distributions, Nat. Commun., 6, 1–8, https://doi.org/10.1038/ncomms9221, 2015.
O'Brien, C. J., Peloquin, J. A., Vogt, M., Heinle, M., Gruber, N., Ajani, P., Andruleit, H., Arístegui, J., Beaufort, L., Estrada, M., Karentz, D., Kopczyńska, E., Lee, R., Poulton, A. J., Pritchard, T., and Widdicombe, C.: Global marine plankton functional type biomass distributions: coccolithophores, Earth Syst. Sci. Data, 5, 259–276, https://doi.org/10.5194/essd-5-259-2013, 2013.
O'Brien, C. J., Vogt, M., and Gruber, N.: Global coccolithophore diversity: Drivers and future change, Prog. Oceanogr., 140, 27–42, https://doi.org/10.1016/j.pocean.2015.10.003, 2016.
Phillips, S. J., Dudík, M., Elith, J., Graham, C. H., Lehmann, A., Leathwick, J., and Ferrier, S.: Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data, Ecol. Appl., 19, 181–197, https://doi.org/10.1890/07-2153.1, 2009.
Provoost, P. and Bosch, S.: robis: R client for the OBIS API. R package version 0.1.5, 2015.
Richardson, A. J., Walne, A. W., John, A. W. G., Jonas, T. D., Lindley, J. A., Sims, D. W., Stevens, D., and Witt, M.: Using continuous plankton recorder data, Prog. Oceanogr., 68, 27–74, https://doi.org/10.1016/j.pocean.2005.09.011, 2006.
Righetti, D., Vogt, M., Zimmermann, N. E., and Gruber, N.: PHYTOBASE: A global synthesis of open ocean phytoplankton occurrences, Pangaea, https://doi.org/10.1594/PANGAEA.904397, 2019a.
Righetti, D., Vogt, M., Gruber, N., Psomas, A., and Zimmermann, N. E.: Global pattern of phytoplankton diversity driven by temperature and environmental variability, Sci. Adv., 5, eaau6253, https://doi.org/10.1126/sciadv.aau6253, 2019b.
Rodríguez-Ramos, T., Marañón, E., and Cermeño, P.: Marine nano- and microphytoplankton diversity: redrawing global patterns from sampling-standardized data, Glob. Ecol. Biogeogr., 24, 527–538, https://doi.org/10.1111/geb.12274, 2015.
Rombouts, I., Beaugrand, G., Ibañez, F., Gasparini, S., Chiba, S., and Legendre, L.: A multivariate approach to large-scale variation in marine planktonic copepod diversity and its environmental correlates, Limnol. Oceanogr., 55, 2219–2229, https://doi.org/10.4319/lo.2010.55.5.2219, 2010.
Sal, S., López-Urrutia, Á., Irigoien, X., Harbour, D. S., and Harris, R. P.: Marine microplankton diversity database, Ecology, 94, 1658–1658, https://doi.org/10.1890/13-0236.1, 2013.
Ser-Giacomi, E., Zinger, L., Malviya, S., De Vargas, C., Karsenti, E., Bowler, C., and De Monte, S.: Ubiquitous abundance distribution of non-dominant plankton across the global ocean, Nat. Ecol. Evol., 2, 1243–1249, https://doi.org/10.1038/s41559-018-0587-2, 2018.
Sogin, M. L., Morrison, H. G., Huber, J. A., Welch, D. M., Huse, S. M., Neal, P. R., Arrieta, J. M., and Herndl, G. J.: Microbial diversity in the deep sea and the underexplored “rare biosphere,” P. Natl. Acad. Sci. USA, 103, 12115–12120, https://doi.org/10.1073/pnas.0605127103, 2006.
Sournia, A., Chrdtiennot-Dinet, M.-J., and Ricard, M.: Marine phytoplankton: how many species in the world ocean?, J. Plankton Res., 13, 1093–1099, https://doi.org/10.1093/plankt/13.5.1093, 1991.
Sunagawa, S., Coelho, L. P., Chaffron, S., Kultima, J. R., Labadie, K., Salazar, G., Djahanschiri, B., Zeller, G., Mende, D. R., Alberti, A., Cornejo-Castillo, F. M., Costea, P. I., Cruaud, C., D'Ovidio, F., Engelen, S., Ferrera, I., Gasol, J. M., Guidi, L., Hildebrand, F., Kokoszka, F., Lepoivre, C., Lima-Mendez, G., Poulain, J., Poulos, B. T., Royo-Llonch, M., Sarmento, H., Vieira-Silva, S., Dimier, C., Picheral, M., Searson, S., Kandels-Lewis, S., Bowler, C., de Vargas, C., Gorsky, G., Grimsley, N., Hingamp, P., Iudicone, D., Jaillon, O., Not, F., Ogata, H., Pesant, S., Speich, S., Stemmann, L., Sullivan, M. B., Weissenbach, J., Wincker, P., Karsenti, E., Raes, J., Acinas, S. G., Bork, P., Boss, E., Bowler, C., Follows, M., Karp-Boss, L., Krzic, U., Reynaud, E. G., Sardet, C., Sieracki, M., and Velayoudon, D.: Structure and function of the global ocean microbiome, Science, 348, 1261359–1261359, https://doi.org/10.1126/science.1261359, 2015.
Thompson, G. G. and Withers, P. C.: Effect of species richness and relative abundance on the shape of the species accumulation curve, Austral Ecol., 28, 355–360, https://doi.org/10.1046/j.1442-9993.2003.01294.x, 2003.
Tittensor, D. P., Mora, C., Jetz, W., Lotze, H. K., Ricard, D., Berghe, E. V., and Worm, B.: Global patterns and predictors of marine biodiversity across taxa, Nature, 466, 1098–1101, https://doi.org/10.1038/nature09329, 2010.
Turland, N. J., Wiersema, J. H., Barrie, F. R., Greuter, W., Hawksworth, D. L., Herendeen, P. S., Knapp, S., Kusber, W.-H., Li, D.-Z., Marhold, K., May, T. W., McNeill, J., Monro, A. M., Prado, J., Price, M. J., and Smith, G. F. (Eds.): International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017, Regnum Vegetabile, Vol. 159, 1–253, Glashütten: Koeltz Botanical Books, https://doi.org/10.12705/Code.2018, 2018.
Utermöhl, H.: Zur Vervollkommnung der quantitativen Phytoplankton-Methodik, SIL Commun. 1953–1996, 9, 1–38, https://doi.org/10.1080/05384680.1958.11904091, 1958.
Villar, E., Farrant, G. K., Follows, M., Garczarek, L., Speich, S., Audic, S., Bittner, L., Blanke, B., Brum, J. R., Brunet, C., Casotti, R., Chase, A., Dolan, J. R., D'Ortenzio, F., Gattuso, J.-P., Grima, N., Guidi, L., Hill, C. N., Jahn, O., Jamet, J.-L., Le Goff, H., Lepoivre, C., Malviya, S., Pelletier, E., Romagnan, J.-B., Roux, S., Santini, S., Scalco, E., Schwenck, S. M., Tanaka, A., Testor, P., Vannier, T., Vincent, F., Zingone, A., Dimier, C., Picheral, M., Searson, S., Kandels-Lewis, S., Acinas, S. G., Bork, P., Boss, E., de Vargas, C., Gorsky, G., Ogata, H., Pesant, S., Sullivan, M. B., Sunagawa, S., Wincker, P., Karsenti, E., Bowler, C., Not, F., Hingamp, P., and Iudicone, D.: Environmental characteristics of Agulhas rings affect interocean plankton transport, Science, 348, 1261447–1261447, https://doi.org/10.1126/science.1261447, 2015.
Vogt, M., O'Brien, C., Peloquin, J., Schoemann, V., Breton, E., Estrada, M., Gibson, J., Karentz, D., Van Leeuwe, M. A., Stefels, J., Widdicombe, C., and Peperzak, L.: Global marine plankton functional type biomass distributions: Phaeocystis spp., Earth Syst. Sci. Data, 4, 107–120, https://doi.org/10.5194/essd-4-107-2012, 2012.
Wallace, D. W. R.: Chapter 6.3 Storage and transport of excess CO2 in the oceans: The JGOFS/WOCE global CO2 survey, in Eos, Transactions American Geophysical Union, Vol. 82, 489–521, 2001.
Wickham, H. and Chang, W.: Devtools: Tools to make developing R packages easier. R package version 1.12.0, 2015.
Woolley, S. N. C., Tittensor, D. P., Dunstan, P. K., Guillera-Arroita, G., Lahoz-Monfort, J. J., Wintle, B. A., Worm, B., and O'Hara, T. D.: Deep-sea diversity patterns are shaped by energy availability, Nature, 533, 393–396, https://doi.org/10.1038/nature17937, 2016.
Worm, B., Sandow, M., Oschlies, A., Lotze, H. K., and Myers, R. A.: Global patterns of predator diversity in the open oceans, Science, 309, 1365–1369, https://doi.org/10.1126/science.1113399, 2005.
Zimmermann, N. E. and Guisan, A.: Predictive habitat distribution models in ecology, Ecol. Modell., 135, 147–186, https://doi.org/10.1016/S0304-3800(00)00354-9, 2000.
Zweng, M. M., Reagan, J. R., Antonov, J. I., Locarnini, R. A., Mishonov, A. V., Boyer, T. P., Garcia, H. E., Baranova, O. K., Johnson, D. R., Seidov, D., and Biddle, M. M.: World Ocean Atlas 2013, Vol. 2, Salinity, edited by: Levitus, S. and Mishonov, A., NOAA Atlas NESDIS 74, 39 pp., 2013.