the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
The Arctic Traits Database – a repository of Arctic benthic invertebrate traits
Sarah Faulwetter
The recently increased interest in marine trait-based studies highlights one general demand – the access to standardized, reference-based trait information. This demand holds especially true for polar regions, where the gathering of ecological information is still challenging. The Arctic Traits Database is a freely accessible online repository (https://doi.org/10.25365/phaidra.49; https://www.univie.ac.at/arctictraits, last access: 20 February 2019) that fulfils these requests for one important component of polar marine life, the Arctic benthic macroinvertebrates. It accounts for (1) obligate traceability of information (every entry is linked to at least one source), (2) exchangeability among trait platforms (use of most common download formats), (3) standardization (use of most common terminology and coding scheme) and (4) user-friendliness (granted by an intuitive web interface and rapid and easy download options, for the first time including the option to download a fuzzy coded trait matrix). The combination of these aspects makes the Arctic Traits Database the currently most sophisticated online accessible trait platform in (not only) marine ecology and a role model for prospective databases of other marine compartments or other (also non-marine) ecosystems. At present the database covers 19 traits (80 trait categories) and holds altogether 14 242 trait entries for 1911 macro- and megabenthic taxa. Thus, the Arctic Traits Database will foster and facilitate trait-based approaches in polar regions in the future and increase our ecological understanding of this rapidly changing system.
- Article
(1877 KB) - Full-text XML
-
Supplement
(1096 KB) - BibTeX
- EndNote
The interest in trait-based approaches – i.e., such that consider the life history, morphological, physiological and behavioral characteristics (i.e., traits) of species – in the marine realm has been growing tremendously in the last decades (reviewed in Degen et al., 2018) (Fig. 1). Reasons for the increasing popularity of these approaches are that they offer a variety of additional options to solely species-based methods: traits can be analyzed across wide geographical ranges and across species pools (Bernhardt-Römermann et al., 2011), and they can be used to calculate a variety of functional diversity indices (Schleuter et al., 2010), to estimate functional redundancy (Darr et al., 2014) or as indicators of ecosystem functioning (Bremner et al., 2006). Given the rapid changes we observe in many marine regions of the world, and especially in the Arctic Ocean (Wassmann et al., 2011), the potential to indicate vulnerability to climate change and biodiversity loss or to estimate climate change effects on ecosystem functions is another inherent advantage of trait-based approaches (Foden et al., 2013; Hewitt et al., 2016).
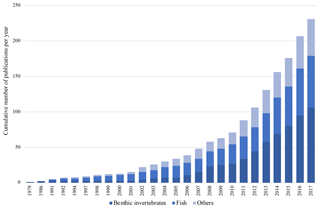
Figure 1Cumulative number of marine trait-based studies based on the literature review of 233 studies from the marine realm by Degen et al. (2018).
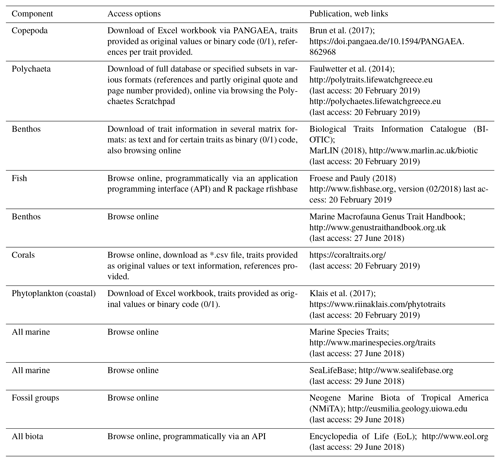
Although the methodical diversity and complexity of trait-based approaches have broadened in the last years (Beauchard et al., 2017; Kleyer et al., 2012), the underlying data are always species traits. Trait information, however, is often not easy to find, and its collation requires a time- and labor-intensive survey of literature, databases, field data, and expert knowledge. This holds especially true for the polar regions, as ecological information for many polar marine taxa is still scarce, and only few publications supplement traceable resources of trait information (e.g., Kokarev et al., 2017). An additional obstacle is that existing trait repositories focus mainly on species from temperate regions. The increasing variability in terminology that surrounds traits is another challenge, and recent publications stress the importance of standardization in order to facilitate meta-analyses and comparison of results (Costello et al., 2015; Degen et al., 2018). Several online accessible trait databases specialize in specific taxonomic groups such as fish, polychaetes or copepods, while others cover a wider part of the marine community (Table 1). The number of traits included and the form of access varies considerably among the different repositories. The database for marine copepods (Brun et al., 2017) contains 14 traits, whereas Fishbase (http://www.fishbase.org, last access: 20 February 2019), polytraits (Faulwetter et al., 2014) and BIOTIC (MarLIN, 2018, http://www.marlin.ac.uk/biotic, last access: 20 February 2019) contain more than 40 traits. Some repositories allow only for online browsing, while others enable different forms of download that range from spreadsheets to different matrix formats (Table 1). No traits repository explicitly comprising polar species has existed so far.
Table 1List of marine trait databases or repositories. “Component” indicates the organism group targeted. “Access options” indicates in which forms the data can be accessed. References and web links are provided.
With the Arctic Traits Database presented here we aim to investigate some of the above-mentioned issues for one important compartment of marine life: the Arctic macro- and megabenthic invertebrates. In order to fulfil the communities' demand for standardization and comparability, only those traits and trait categories are included that are most frequently used in topical publications or which are already provided in freely accessible trait databases (Table 1). Regarding download options and traceability, we follow the successful example given in Faulwetter et al. (2014) and provide download of trait data in different tabular formats (i.e., data in columns, once following a database-specific format and once following Darwin Core format) (Wieczorek et al., 2012). The use of these formats guarantees that the included trait information can be easily shared between trait repositories and that the content is fully exploitable both by humans and computers. Every trait code is backed up by at least one reference, and where possible the original quote and page number are provided. In addition to the above-mentioned formats, for the first time trait information has also been made available in a fuzzy-coded and ready-to-use matrix format that can be directly incorporated into appropriate analysis software.
By providing the Arctic Traits Database to the community of benthic ecologists, we aim to provide a sound basis for prospective trait-based approaches in polar regions which will in return aid our overall understanding of these unique and rapidly changing ecosystems.
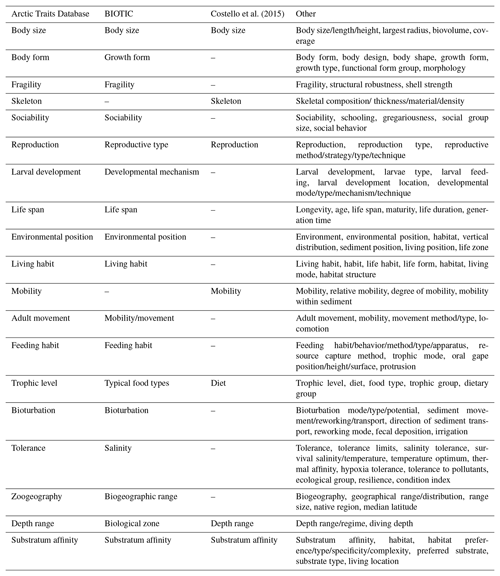
Table 2Trait terminology as used in the Arctic Traits Database, BIOTIC, Costello et al. (2015) and in other marine trait-based studies (i.e., studies reviewed in Degen et al., 2018, list non-exhaustive; see Appendix 1 of Degen et al., 2018 for total trait list and corresponding literature references). Be aware that the Arctic Traits Database and BIOTIC only consider benthic taxa, while Costello et al. (2015) and the studies summarized in “Other” cover all marine groups.
2.1 Taxon data
The current taxa in the database are a subset of the dataset compiled in the frame of the “Arctic Traits Project” (Austrian Science Fund FWF, T801-B29), with a focus on pan-Arctic benthic invertebrate macro- and megafauna. This dataset comprises species lists from published studies of collaborators (Blanchard et al., 2013a, b; Grebmeier et al., 2015) but also from sampling campaigns that are so far unpublished (e.g., field courses of the University Center in Svalbard, UNIS, 2007–2017). The regional coverage currently comprises the Chukchi Sea and the Svalbard area. At present, mainly species in the macrofauna size class have been uploaded.
2.2 Trait data
Here, we consider 19 traits and 80 trait categories that reflect the morphology, life history and the behavior of Arctic benthic invertebrates (Table 3). All traits are in categorical format; i.e., they belong to one out of up to six clearly defined trait categories (see Table 3). The three continuous traits included (body size, life span and depth distribution) are converted into categories, but the associated text information also assures accessibility to users in their original numerical or continuous format.
Table 3Detailed information on the 19 biological traits currently included in the Arctic Traits Database, clustered into morphology traits (5), life history traits (3) and behavioral traits (11). For every trait and its categories, the definition as used in the Arctic Traits Database is given. Abbreviations of each category are given (e.g., S1, S2) as these are used in files downloaded from the website. The relation of the respective trait to benthic ecosystem functions or responses (i.e., its role as effect or response trait) is given via specific examples, and underlying literature sources are displayed.
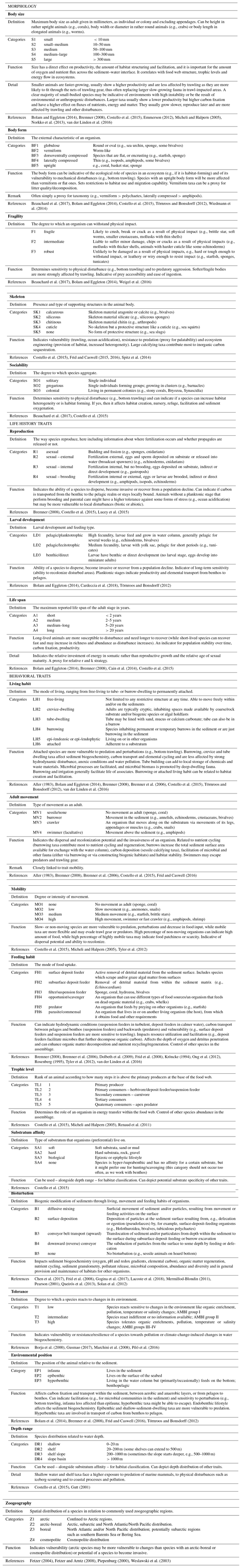
The choice of which traits to include in the database is based on the following considerations: (1) trait information should be available for and applicable to all benthic taxa (Costello et al., 2015); (2) traits used in previous studies and databases should be favored to enable comparisons across studies (Degen et al., 2018); and (3) the traits should be usable across a wide geographical area (Bremner et al., 2006). In order to fulfil this last precondition, the trait body size is provided as the “maximum body size as adult” (see also Table 3). While clearly a trade-off in regard to the detection of intraspecific plasticity, it enables the use of this trait across large spatial scales.
Recent trait-based studies emphasize the importance of standardized traits and trait terminology to ensure that data can be integrated more easily in the future (Costello et al., 2015; Degen et al., 2018; Faulwetter et al., 2014). To meet these requirements of the scientific community, the Arctic Traits Database includes 7 of the 10 traits prioritized in Costello et al. (2015): depth range, substratum affinity, mobility, skeleton, diet, body size and reproduction (Table 3). The remaining three traits emphasized in Costello et al. (2015) – taxonomic identity, environment and geography – are not included. For taxonomic traits, every species in the database is bidirectionally deep linked (i.e., connected via a hyperlink) to the World Register of Marine Species (WoRMS Editorial Board, 2017; http://www.marinespecies.org/, last access: 20 February 2019). For more detailed biogeographic information we refer users to the Global Biodiversity Information System (GBIF; http://www.gbif.org/, last access: 29 June 2018) or the Ocean Biogeographic Information System (OBIS; http://www.iobis.org, last access: 27 June 2018). We do include, however, the trait “zoogeography”, which enables a differentiation between typical arctic and boreal or cosmopolitan taxa. Of the 19 traits used here, 17 are also identical to those used by the BIOTIC database (MarLIN 2006, Table 1), one of the most comprehensive databases on biological traits of marine organisms. BIOTIC also includes the trait “salinity”. We cover salinity preferences within the trait “tolerance”, which also accounts for temperature and pollution tolerance (see Table 3 for details). Traits we include in addition are “skeleton”, and “mobility” (i.e., the relative degree of movement). Although physiological traits are of high interest in trait-based studies, we do not include them as they are not easily retrieved for many (arctic) benthic taxa (one of the preconditions for inclusion in the database as stated above). In addition, physiological traits (e.g., growth rate, respiration rate, ingestion rate) depend on body mass and temperature (Brown et al., 2004), which can vary tremendously among Arctic regions, contradicting the consideration that the trait information provided should be usable across a wide geographical area.
One common approach to the use of traits is as indicators of ecosystem functions (effect traits) or of changes in the environment (response traits) (Hooper et al., 2005). An overview of how each of the 19 traits that are currently included in the database may relate to ecosystem functions or respond to environmental changes or pressures is given in Table 3.
2.3 Sources of trait information
Sources of trait information are research papers, books, databases and online repositories (Table 1) but also grey literature such as cruise reports. Trait information can also result from on-site measurements (e.g., for the trait body size) or personal observations or be transmitted via communication with experts for a specific taxonomic group. In any case, the source is indicated as precisely as possible, for published literature with a complete reference and DOI (if available), and in the case of expert communication, the name and contact details of the respective expert are given. Wherever possible the original quote from literature and page numbers are given to ensure the traceability of the provided trait information. Although literature sources targeting the Arctic are used preferably (and for exclusively Arctic species are the only option), we do not restrict source information for arctic-boreal or cosmopolite taxa to stem from Arctic regions. This bears the risk that the assigned trait information is not accurate, as polar taxa might differ in their expression of certain traits from their relatives at lower latitudes (Degen et al., 2018). However, this is an issue that is not resolved for now, as trait information from the high latitudes is often scarce, and the user is recommended to consider the source of trait information when interpreting results.
2.4 Fuzzy coding of traits
The fuzzy coding procedure indicates to what extent a taxon exhibits each trait category (Chevenet et al., 1994). This method has the advantage that it enables us to analyze diverse kinds of biological information derived from a variety of sources (as those included in the Arctic Traits Database; see Sect. 2.3), and that also intermediate scenarios (i.e., when a taxon does not clearly fall into one category or the other) can be accounted for (Chevenet et al., 1994). We use the 0–3 coding scheme (details in Table 4 above) as it is the most widely used (which facilitates comparisons and exchange of trait information) and provides a compromise between binary codes and many graduations that are not clearly delineated (Degen et al., 2018).
Table 5Two coding examples for the trait “Feeding habit”, which has six trait categories (FH1–FH6; see also Table 3). Species 1 is a surface deposit feeder but can switch from facultative to suspension feeding, while species 2 is an exclusive suspension feeder.
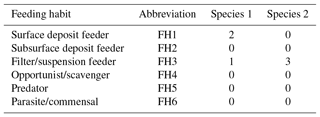
While the coding might be pretty straightforward for some traits and taxa, in some cases a decision might not be drawn so easily. As one of the clearer cases, we point out the coding of the trait “body size” for the starfish Crossaster papposus. A literature reference states that the body size can range “up to 340 mm in diameter” (Hayward and Ryland, 2012, p. 668). This size fits into the category “large” (S5, >300 mm); thus the taxon is coded “3” for this size class, and “0” for all other categories (S1–S4). The trait “mobility” is trickier. A literature reference (Himmelman and Dutil, 1991, p. 68) states the following: “Crossaster papposus and Solaster endeca are highly mobile; large individuals can cover distances of more than 5 meters in 12 hours”. Here we have to keep in mind that the particular reference frame in this publication is subtidal sea stars in the northern Gulf of St. Lawrence (west Atlantic). The reference of the Arctic Traits Database however is the entire community of benthic invertebrates, and the trait category “high mobility” is defined here for taxa which are “swimmers or fast crawlers”, such as some amphipods and shrimp (see Table 2). Accordingly, the correct coding for C. papposus in the reference system of the Arctic Traits Database is the category “medium” mobility (MO3). Users of the Arctic Traits Database should bear this reference system in mind when only downloading the fuzzy coded trait data and aiming to apply it to another reference system. But as the detailed literature quote that leads to the coding of a trait is always provided (see Sect. 2.3), the trait information can easily be adjusted by the user.
Table 6This is how the above example would appear in the matrix downloaded from the Arctic Traits Database. In the download matrix format species are rows, trait categories are columns and the fuzzy codes are the values. Due to the database structure zero codes (“0”) are only displayed when they are backed up by a specific reference (e.g., for the trait category LH3/tube dwelling: “No species within the family Polynoidae is tubiculous”).
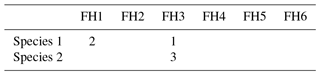
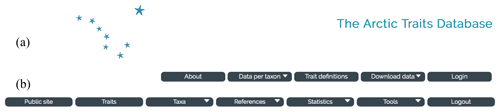
Figure 2Screenshots of the start page of the Arctic Traits Database. Toolbar of the public page with the login button for the registered user (a) and toolbar in the area for registered users (b).
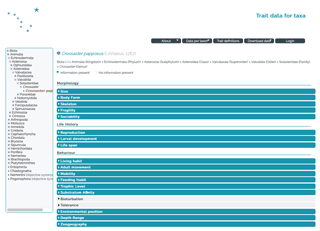
Figure 3Screenshot of the taxon page of the asteroid Crossaster papposus selected from the classification tree on the left.
There will always be a certain degree of subjectivity related to the fuzzy coding procedure. To find out how strong the coding might differ among scientists, a small experiment was performed at the Arctic Traits Workshop in Vienna (December 2016) (Degen et al., 2018). Participants coded 27 trait categories of three common Arctic benthic species and found the final trait matrices to be to 83 % identical. We are confident that the sophisticated structure of the Arctic Traits Database (see Sect. 3) and the information and instructions provided will support a more consistent coding of benthic traits in the future.
In order to collect trait information and to disseminate it among users, a web-based database was created. The database features a public interface (Sect. 3.1) and an entry interface that is only accessible for registered collaborators (Supplement). The public interface (Fig. 2a) allows the traits and references to be browsed online (“Data per taxon” in the top menu bar), background information to be viewed (“About” and “Trait definitions”) and either the entire species, trait and literature information or specified subsets to be downloaded in several formats (“Download data”) (see Sect. 3.1). Registered collaborators – i.e., those users that actively contribute trait information to the Arctic Traits Database – can access the interactive part of the database via the login button on the public page (Fig. 2a). This access offers additional options (Fig. 2b): browsing the existing information also by trait (“Traits” in the top menu bar), uploading new taxa, trait and source information or adding trait information, references and comments to already existing taxa in the database (“Taxa”). As several users can work on the same taxa, a flagging system is used to highlight and discuss potentially conflicting sources and opinions.
Table 7List of fields returned by the Arctic Traits Database when “Data as columns” (*.csv) is chosen as an export option from the download section.
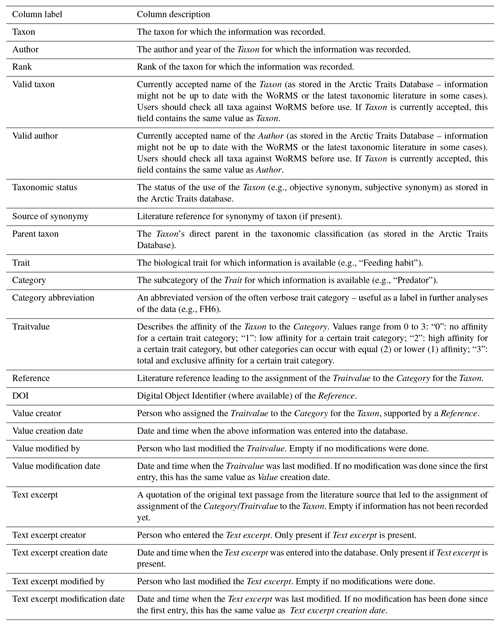
Table 8List of fields returned by the Arctic Traits Database when “Darwin Core” is chosen as an export option from the download section. Darwin Core does not provide the same granularity as the “Data as columns” format. The output file consequently contains fewer details.
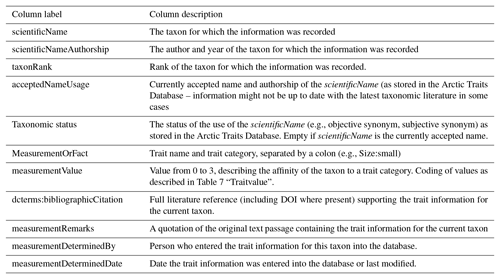
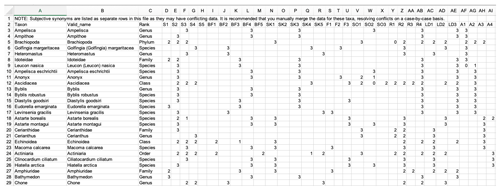
Figure 5A screenshot from the fuzzy coded trait matrix returned by the Arctic Traits Database when the “Data in matrix format” is chosen as export option from the download section. Species are rows (“Valid_name” refers to the currently accepted taxonomy in WoRMS), and abbreviated trait categories are columns. For abbreviations of trait categories, see Table 3. Due to the database structure, zero codes (“0”) are not displayed (see Table 6).
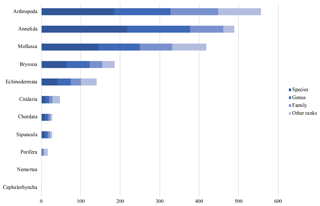
Figure 6Taxonomic data coverage. “Other ranks” include higher taxonomic levels and intermediate ranks.
The “References”, “Statistics” and “Tools” sections are equally only accessible for registered users (Fig. 2b; Supplement). Every scientist working in the field of Arctic benthic ecology aiming to share trait information can become a registered user by getting in touch with the editor and retrieving a user login. Credit to the registered collaborators is given in the “About” section on the public site and also on taxon pages after each trait entry they conduct. A detailed manual for registered users is provided in the Supplement to this publication or can alternatively be accessed via the public web interface (“About”). Collaborators who want to share trait information without registering on the database can alternatively be provided with an upload template xls.
3.1 Public access and download options
The public access enables the database to be browsed online and the complete set of data as well as the bibliography or specified subsets to be downloaded. Taxon traits can be visually inspected online via the “Data per taxon” button from the top menu bar and “Browse taxa” or “Search taxa”. Taxa can be browsed and selected via the taxonomic tree, as indicated for the asteroid Crossaster papposus in Fig. 3. Alternatively, the “Search taxa” panel allows a specific taxon to be typed in and searched.
The completeness of trait information can be inspected via “Data completeness” (Fig. 4), equally accessible via “Data per taxon” on the top menu bar. This option shows an alphabetic list of all taxa in the database for which trait information is available. The bar on the right side indicates the information coverage for each taxon and trait; the blue color indicates that trait information is present.
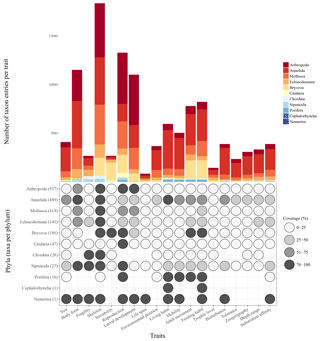
Figure 7Scheme visualizing the taxon entries per trait (bar chart), the number of taxa per phylum (brackets) and the data coverage per trait per phylum (dot plot).
The download section can be accessed via the “Download data” button on the top menu bar (Figs. 2a, 3, 4). Download is enabled in three different computer readable formats: (1) as data in columns (*.csv) (Table 7), (2) in Darwin Core format (Table 8) and (3) as a fuzzy coded trait matrix, which some users might prefer (see Sect. 2.4 and Fig. 5). Also, the entire bibliography is available for download. Before the download commences the user is asked whether to download (a) all data in the database, (b) only data for an uploaded list of taxon names, (c) only data for an uploaded list of AphiaIDs, or (d) only the data selected from a classification tree. In the last option, entire phyla or sub-groups can be easily selected from the tree. By default, all 19 traits are exported, but if the user is interested only in one or a few specific traits, the option to select these from the total list of 19 traits is available. As the fuzzy coded trait matrix (download option 3) contains only the fuzzy codes per trait category but no literature sources, we recommend also downloading the data in columns (download option 1) for the same taxa, for which the detailed source per species and trait category is included. Details on the structure of the first two download options are given above in Tables 7 and 8. A screenshot from a downloaded fuzzy coded trait matrix is shown in Fig. 5. The database can also be accessed programmatically via a REST API (documented at https://www.univie.ac.at/arctictraits/download-api, last access: 20 February 2019).
3.2 Database specification
The website runs on an Apache 2.2. server, and the database is implemented in MySQL 5. PHP 5 is used as the scripting language. Web technologies used are HTML4, CSS and JavaScript/Jquery. A code package to create such a web-based trait database including a README file with instructions for installation is provided at figshare; https://doi.org/10.6084/m9.figshare.7491869.
4.1 Taxonomic data coverage
At present, the database contains 1911 Arctic marine benthic invertebrate taxa. Thereof 686 are on species level, 516 are on genus level and 274 are on family level. The remaining 435 taxa are higher taxonomic levels or intermediate ranks. The largest taxonomic group in the database at present stage are the Arthropoda with 557 taxa (186 entries on species level), followed by the Annelida with 489 taxa (218 entries on species level) and the Mollusca with 418 taxa (146 entries on species level) (Fig. 6).
4.2 Trait data coverage
At present, the database contains 19 traits and 80 trait categories, with currently 14 242 entries of trait information in total. The trait for which most entries exist is “Skeleton” (1837 entries), followed by “Reproduction” (1328 entries) and “Body form” (1151 entries) (Fig. 7). The phylum with most entries is the Annelida (6130 entries, 43 %), followed by Arthropoda (2968 entries, 21 %) and Mollusca (2177 entries, 15 %). Regarding the taxonomic level, most trait information was added on the species level (48 %), less on the genus (25 %) and family level (17 %).
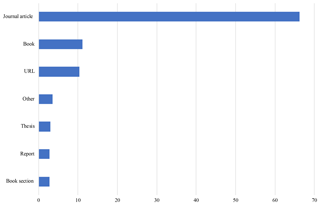
4.3 Bibliography
The Arctic Traits Database currently includes 394 sources of trait information. Thereof 66 % are scientific papers, 11 % are books, 10 % are web pages and 4 % are expert communications and personal observations (“Other”). Theses, book sections and reports each make up around 3 %. Most sources were used for the phyla Echinodermata and Annelida (33 % each), followed by Arthropoda (29 %).
Although the Arctic Traits Database is still growing as new taxa and trait information are added, certain trends in data completeness or scarceness, respectively, have become apparent (Fig. 7). Thus, the database is not only a valuable tool for collecting and providing information but also for pointing out in which areas more research might be needed. Regarding the 19 traits included at the present stage, it shows that our knowledge on, e.g., the life span of many Arctic benthic species is still limited (information only for <5 % of species). This lack of data on species longevity is astonishing, as polar taxa are traditionally depicted as slow-growing and long-lived compared to their relatives from lower latitudes. Accordingly, one might have expected that more studies and measurements are available for a variety of Arctic taxa, which is not the case for many groups. Other traits that are currently underrepresented are trophic level (<8 %) and tolerance (<13 %).
Regarding our interest in identifying knowledge gaps, a special strength of the database is the implemented flagging system (described in detail in the Supplement). As registered users continue to upload trait information, more “conflicts” – i.e., cases in which the sources or observations added by different users point towards different trait categories – may also arise. Such cases are then indicated by a red flag and can be easily filtered. Monitoring and statistical evaluation of these cases will grant important information on where conflicts exist and for which taxa or traits future research is needed. Such evaluation will also aid in identifying which traits are more robust (i.e., are never flagged) and which show a higher plasticity (frequent flagging). This kind of information is of tremendous value as it can aid the choice as to which traits to include in prospective trait-based studies. Apart from clearly diverging source information, different levels of experience or customs in fuzzy coding might also lead to red flags in the system. Here the editorial team will take care of consistency by solving the conflicts according to the database standard and through that also fostering a standardized way of coding within the community. In addition, repetitively occurring discrepancies in the coding of certain traits might also point towards a need for revision of these trait categories or their definitions, or maybe even the adding of a new trait, in that way improving the quality of the database.
In addition to the knowledge gaps surrounding certain traits discussed above, the data coverage among taxonomic groups also varies considerable (Fig. 7). This potentially mirrors the sampling design of the underlying datasets. Some taxonomic groups such as the polychaetes clearly dominate many benthic soft-bottom communities, while other taxa such as the shrimp/Caridea are highly mobile and might be permanently undersampled with sampling gear like grabs, box corers or bottom trawls (Eleftheriou and McIntyre, 2007). This points toward the need of also including datasets derived from video and still image analysis in the future development of the database. These methods – despite certain disadvantages (discussed in Degen et al., 2018, their supplementary material, file 3) – have the great benefit that traits of hard-bottom communities can also be analyzed, ecosystems which are at present underrepresented in the Arctic Traits Database.
The Arctic Traits Database is hosted at the University of Vienna (Austria) and can be accessed via https://www.univie.ac.at/arctictraits/ (last access: 20 February 2019) (https://doi.org/10.25365/phaidra.49; Degen and Faulwetter, 2018). A code package to create a web-based trait database including a README file with instructions for installation is provided at figshare; https://doi.org/10.6084/m9.figshare.7491869 (Faulwetter and Degen, 2018).
The Arctic Traits Database provides an easy accessible and sound knowledge base of traits of Arctic benthic invertebrates and will thus facilitate prospective trait-based studies for a variety of benthic ecologists at all career stages. Its sophisticated structure accounts for the most commonly raised demands for contemporary trait databases: (1) obligate traceability of information (every entry is linked to at least one source); (2) exchangeability among platforms (use of most common download formats); (3) standardization (use of most common terminology and coding scheme); and last but not least (4) user-friendliness (granted by an intuitive web interface and rapid and easy download options). The combination of these aspects makes the Arctic Traits Database a cutting-edge tool for (not only) the marine realm and a role model for prospective databases.
The supplement related to this article is available online at: https://doi.org/10.5194/essd-11-301-2019-supplement.
RD designed the project and performed the trait data collection. SF performed database and web page development and design. RD prepared the manuscript with contributions from SF.
The authors declare that they have no conflict of interest.
The authors wish to thank all collaborators that support the Artic Traits
Project, especially Bodil Bluhm, Jackie Grebmeier, Lauren Sutton, Dieter
Piepenburg and Arny Blanchard. This work was supported by the Austrian
Science Fund (FWF; T 801-B29) to Renate
Degen.
Edited by: David Carlson
Reviewed by: two anonymous referees
Aller, R. C.: The importance of the diffusive permeability of animal burrow linings in determining marine sediment chemistry, J. Mar. Res., 41, 299–322, https://doi.org/10.1357/002224083788520225, 1983.
Beauchard, O., Veríssimo, H., Queirós, A. M., and Herman, P. M. J.: The use of multiple biological traits in marine community ecology and its potential in ecological indicator development, Ecol. Indic., 76, 81–96, https://doi.org/10.1016/j.ecolind.2017.01.011, 2017.
Bernhardt-Römermann, M., Gray, A., Vanbergen, A. J., Bergès, L., Bohner, A., Brooker, R. W., Bergès, L., Bohner, A., Brooker, R. W., De Bruyn, L., De Cinti, B., Dirnböck, T., Grandin, U., Hester, A. J., Kanka, R., Klotz, S., Loucougaray, G., Lundin, L., Matteucci, G., Mészáros, I., Oláh, V., Preda, E., Prévosto, B., Pykälä, J., Schmidt, W., Taylor, M. E., Vadineanu, A., Waldmann, T., and Stadler, J.: Functional traits and local environment predict vegetation responses to disturbance: a pan-European multi-site experiment, J. Ecol., 99, 777–787, https://doi.org/10.1111/j.1365-2745.2011.01794.x, 2011.
Blanchard, A. L., Parris, C. L., Knowlton, A. L., and Wade, N. R.: Benthic ecology of the northeastern Chukchi Sea. Part I. Environmental characteristics and macrofaunal community structure, 2008–2010, Cont. Shelf Res., 67, 52–66, https://doi.org/10.1016/j.csr.2013.04.021, 2013a.
Blanchard, A. L., Parris, C. L., Knowlton, A. L., and Wade, N. R.: Benthic ecology of the northeastern Chukchi Sea. Part II. Spatial variation of megafaunal community structure, 2009–2010, Cont. Shelf Res., 67, 67–76, https://doi.org/10.1016/j.csr.2013.04.031, 2013b.
Bolam, S. G. and Eggleton, J. D.: Macrofaunal production and biological traits: Spatial relationships along the UK continental shelf, J. Sea Res., 88, 47–58, https://doi.org/10.1016/j.seares.2014.01.001, 2014.
Borja, A., Franco, J., and Pérez, V.: A Marine Biotic Index to Establish the Ecological Quality of Soft-Bottom Benthos Within European Estuarine and Coastal Environments, Mar. Pollut. Bull., 40, 1100–1114, 2000.
Bremner, J.: Species' traits and ecological functioning in marine conservation and management, J. Exp. Mar. Biol. Ecol., 366, 37–47, https://doi.org/10.1016/j.jembe.2008.07.007, 2008.
Bremner, J., Rogers, S. I., and Frid, C. L. J.: Methods for describing ecological functioning of marine benthic assemblages using biological traits analysis (BTA), Ecol. Indic., 6, 609–622, https://doi.org/10.1016/j.ecolind.2005.08.026, 2006.
Brown, J. H., Gillooly, J. F., Allen, A. P., Van Savage, M., and West, G. B.: Toward a metabolic theory of ecology, Ecology, 85, 1771–1789, 2004.
Brun, P., Payne, M. R., and Kiørboe, T.: A trait database for marine copepods, Earth Syst. Sci. Data, 9, 99–113, https://doi.org/10.5194/essd-9-99-2017, 2017.
Cain, M. L., Bowman, W. D., and Hacker, S. D.: Ecology, 3rd Edn., Sinauer Associates, 2014.
Cardeccia, A., Marchini, A., Occhipinti-Ambrogi, A., Galil, B., Gollasch, S., Minchin, D., Naršcius, A., Olenin, S., and Ojaveer, H.: Assessing biological invasions in European Seas: Biological traits of the most widespread non-indigenous species, Estuar. Coast. Shelf S., 201, 17–28, https://doi.org/10.1016/j.ecss.2016.02.014, 2018.
Chen, X., Andersen, T. J., Morono, Y., Inagaki, F., Jørgensen, B. B., and Lever, M. A.: Bioturbation as a key driver behind the dominance of Bacteria over Archaea in near-surface sediment, Sci. Rep.-UK, 7, 1–14, https://doi.org/10.1038/s41598-017-02295-x, 2017.
Chevenet, F., Dolédec, S., and Chessel, D.: A fuzzy coding approach for the analysis of long-term ecological data, Freshwater Biol., 31, 295–309, https://doi.org/10.1111/j.1365-2427.1994.tb01742.x, 1994.
Costello, M. J., Claus, S., Dekeyzer, S., Vandepitte, L., Tuama, É. Ó., Lear, D., and Tyler-Walters, H.: Biological and ecological traits of marine species, PeerJ, 3, e1201, https://doi.org/10.7717/peerj.1201, 2015.
Darr, A., Gogina, M., and Zettler, M. L.: Functional changes in benthic communities along a salinity gradient- a western Baltic case study, J. Sea Res., 85, 315–324, https://doi.org/10.1016/j.seares.2013.06.003, 2014.
Degen, R. and Faulwetter, S.: The Arctic Traits Database, https://doi.org/10.25365/phaidra.49, 2018.
Degen, R., Aune, M., Bluhm, B. A., Cassidy, C., Kedra, M., Kraan, C., Vandepitte, L., Wlodarska-Kowalczuk, M., Zhulay, I., Albano, P. G., Bremner, J., Grebmeier, J. M., Link, H., Morata, N., Nordström, M. C., Shojaei, M. G., Sutton, L., and Zuschin, M.: Trait-based approaches in rapidly changing ecosystems: A roadmap to the future polar oceans, Ecol. Indic., 91, 722–736, https://doi.org/10.1016/j.ecolind.2018.04.050, 2018.
Dolbeth, M., Teixeira, H., Marques, J. C., and Pardal, M. Â.: Feeding guild composition of a macrobenthic subtidal community along a depth gradient, Sci. Mar., 73, 225–237, https://doi.org/10.3989/scimar.2009.73n2225, 2009.
Eleftheriou, A. and McIntyre, A.: Methods for the Study of Marine Benthos, 3rd Edn., Blackwell Science Ltd., 2007.
Emmerson, M. C.: The importance of body size, abundance, and food-web structure for ecosystem functioning, in: Marine Biodiversity and Ecosystem Functioning: Frameworks, methodologies, and integration, edited by: Solan, M., Aspden, R. J., and Paterson, D. M., Oxford University Press, Oxford, 240 pp., 2012.
Faulwetter, S. and Degen, R.: Code for a web-based biological traits database, https://doi.org/10.6084/m9.figshare.7491869, 2018.
Faulwetter, S., Markantonatou, V., Pavloudi, C., Papageorgiou, N., Keklikoglou, K., Chatzinikolaou, E., Pafilis, E., Chatzigeorgiou, G., Vasileiadou, K., Dailianis, T., Fanini, L., Koulouri, P., and Arvanitidis, C.: Polytraits: A database on biological traits of marine polychaetes, Biodivers. Data J., 2, e1024, https://doi.org/10.3897/BDJ.2.e1024, 2014.
Fetzer, I.: Reproduction strategies and distribution of larvae and juveniles of benthic soft-bottom invertebrates in the Kara Sea (Russian Arctic), PhD thesis, University of Bremen, Germany, 242 pp., 2004.
Fetzer, I. and Arntz, W. E.: Reproductive strategies of benthic invertebrates in the Kara Sea (Russian Arctic): Adaptation of reproduction modes to cold water, Mar. Ecol. Prog. Ser., 356, 189–202, https://doi.org/10.3354/meps07271, 2008.
Foden, W. B., Butchart, S. H. M., Stuart, S. N., Vié, J. C., Akçakaya, H. R., Angulo, A., DeVantier, L. M., Gutsche, A., Turak, E., Cao, L., Donner, S. D., Katariya, V., Bernard, R., Holland, R. A., Hughes, A. F., O'Hanlon, S. E., Garnett, S. T., Sekercioglu, Ç. H., and Mace, G. M.: Identifying the world's most climate change vulnerable species: a systematic trait-based assessment of all birds, amphibians and corals, edited by: Lavergne, S., PLoS One, 8, e65427, https://doi.org/10.1371/journal.pone.0065427, 2013.
Frid, C. L. J. and Caswell, B. A.: Is long-term ecological functioning stable: The case of the marine benthos?, J. Sea Res., 98, 15–23, https://doi.org/10.1016/j.seares.2014.08.003, 2015.
Frid, C. L. J. and Caswell, B. A.: Does ecological redundancy maintain functioning of marine benthos on centennial to millennial time scales?, Mar. Ecol., 37, 392–410, https://doi.org/10.1111/maec.12297, 2016.
Frid, C. L. J., Paramor, O. A. L., Brockington, S., and Bremner, J.: Incorporating ecological functioning into the designation and management of marine protected areas, edited by: Davenport, J., Burnell, G., Cross, T., Emmerson, M., McAllen, R., Ramsay, R., and Rogan, E., Hydrobiologia, 606, 69–79, https://doi.org/10.1007/978-1-4020-8808-7_7, 2008.
Froese, R. and Pauly, D. (Eds.): FishBase, World Wide Web electronic publication, available at: http://www.fishbase.org (last access: 20 February 2019), version (02/2018), 2018.
Gogina, M., Morys, C., Forster, S., Gräwe, U., Friedland, R., and Zettler, M. L.: Towards benthic ecosystem functioning maps: Quantifying bioturbation potential in the German part of the Baltic Sea, Ecol. Indic., 73, 574–588, https://doi.org/10.1016/j.ecolind.2016.10.025, 2017.
Grebmeier, J., Bluhm, B., Cooper, L., Denisenko, S., Iken, K., Kedra, M., and Serratos, C.: Time-Series Benthic Community Composition and Biomass and Associated Environmental Characteristics in the Chukchi Sea During the RUSALCA 2004–2012 Program, Oceanography, 28, 116–133, https://doi.org/10.5670/oceanog.2015.61, 2015.
Gusmao, J. B.: Sediments and Functional Traits?: Applying a Functional Trait Approach To Assess Marine Macrobenthic Function, Univeridade Federal do Parana, 2017.
Gutt, J.: On the direct impact of ice on marine benthic communities, a review, Polar Biol., 24, 553–564, https://doi.org/10.1007/s003000100262, 2001.
Hayward, P. J. and Ryland, J. S. (Eds.): Handbook of the marine fauna of North-West Europe, Oxford University Press, Oxford, New York, Tokyo, 2012.
Hewitt, J. E., Norkko, J., Kauppi, L., Villnäs, A., Norkko, A., and Peters, D. P. C.: Species and functional trait turnover in response to broad-scale change and an invasive species, Ecosphere, 7, e01289, https://doi.org/10.1002/ecs2.1289, 2016.
Himmelman, J. H. and Dutil, C.: Distribution, population structure and feeding of subtidal seastars in the northern Gulf of St. Lawrence, Mar. Ecol. Prog. Ser., 76, 61–72, https://doi.org/10.3354/meps076061, 1991.
Hooper, D. U., Chapin, F. S., Ewel, J. J., Hector, A., Inchausti, P., Lavorel, S., Lawton, J. H., Lodge, D. M., Loreau, M., Naeem, S., Schmid, B., Setälä, H., Symstad, A. J., Vandermeer, J., and Wardle, D. A.: Effects of biodiversity on ecosystem functioning: A consensus of current knowledge, Ecol. Monogr., 75, 3–35, https://doi.org/10.1890/04-0922, 2005.
Klais, R., Norros, V., Lehtinen, S., Tamminen, T., and Olli, K.: Community assembly and drivers of phytoplankton functional structure, edited by: Carrington, E., Funct. Ecol., 31, 760–767, https://doi.org/10.1111/1365-2435.12784, 2017.
Kleyer, M., Dray, S., Bello, F., Lepš, J., Pakeman, R. J., Strauss, B., Thuiller, W., and Lavorel, S.: Assessing species and community functional responses to environmental gradients: Which multivariate methods?, edited by: Wildi, O., J. Veg. Sci., 23, 805–821, https://doi.org/10.1111/j.1654-1103.2012.01402.x, 2012.
Kokarev, V. N., Vedenin, A. A., Basin, A. B., and Azovsky, A. I.: Taxonomic and functional patterns of macrobenthic communities on a high-Arctic shelf: A case study from the Laptev Sea, J. Sea Res., 129, 61–69, https://doi.org/10.1016/j.seares.2017.08.011, 2017.
Kröncke, I.: Macrobenthos composition, abundance and biomass in the Arctic Ocean along a transect between Svalbard and the Makarov Basin, Polar Biol., 14, 519–529, https://doi.org/10.1007/BF00238221, 1994.
Lacoste, É., Piot, A., Archambault, P., McKindsey, C. W., and Nozais, C.: Bioturbation activity of three macrofaunal species and the presence of meiofauna affect the abundance and composition of benthic bacterial communities, Mar. Environ. Res., 136, 62–70, https://doi.org/10.1016/j.marenvres.2018.02.024, 2018.
Lucey, N. M., Lombardi, C., DeMarchi, L., Schulze, A., Gambi, M. C., and Calosi, P.: To brood or not to brood: Are marine invertebrates that protect their offspring more resilient to ocean acidification?, Sci. Rep.-UK, 5, 12009, https://doi.org/10.1038/srep12009, 2015.
Marchini, A., Munari, C., and Mistri, M.: Functions and ecological status of eight Italian lagoons examined using biological traits analysis (BTA), Mar. Pollut. Bull., 56, 1076–1085, https://doi.org/10.1016/j.marpolbul.2008.03.027, 2008.
MarLIN: BIOTIC – Biological Traits Information Catalogue, Marine Life Information Network, Plymouth, Marine Biological Association of the United Kingdom, available at: http://www.marlin.ac.uk/biotic/, last access: 18 April 2018.
Mermillod-Blondin, F.: The functional significance of bioturbation and biodeposition on biogeochemical processes at the water–sediment interface in freshwater and marine ecosystems, J. N. Am. Benthol. Soc., 30, 770–778, https://doi.org/10.1899/10-121.1, 2011.
Micheli, F. and Halpern, B. S.: Low functional redundancy in coastal marine assemblages, Ecol. Lett., 8, 391–400, https://doi.org/10.1111/j.1461-0248.2005.00731.x, 2005.
Norkko, A., Villnäs, A., Norkko, J., Valanko, S., and Pilditch, C.: Size matters: implications of the loss of large individuals for ecosystem function, Sci. Rep.-UK, 3, 2646, https://doi.org/10.1038/srep02646, 2013.
Oug, E., Fleddum, A., Rygg, B., and Olsgard, F.: Biological traits analyses in the study of pollution gradients and ecological functioning of marine soft bottom species assemblages in a fjord ecosystem, J. Exp. Mar. Biol. Ecol., 432, 94–105, available at: http://apps.webofknowledge.com/full_record.do?product=CCC&search_mode=GeneralSearch&qid=7&SID=W29TQeqCuCsBtQBk9hp&page=1&doc=9 (last access: 28 April 2016), 2012.
Pearson, T. H.: Functional group ecology in soft-sediment marine benthos: the role of bioturbation, Oceanogr. Mar. Biol., 39, 78–94, 2001.
Piepenburg, D.: Arctic Brittle Stars (Echinodermata: Ophiuroidea), Oceanogr. Mar. Biol., 38, 189–256, 2000.
Piló, D., Ben-Hamadou, R., Pereira, F., Carriço, A., Pereira, P., Corzo, A., Gaspar, M. B., and Carvalho, S.: How functional traits of estuarine macrobenthic assemblages respond to metal contamination?, Ecol. Indic., 71, 645–659, https://doi.org/10.1016/j.ecolind.2016.07.019, 2016.
The Polychaetes Scratchpad: available at: http://polychaetes.lifewatchgreece.eu/, last access: 27 June 2018
Queirós, A. M., Birchenough, S. N. R., Bremner, J., Godbold, J. A., Parker, R. E., Romero-Ramirez, A., Reiss, H., Solan, M., Somerfield, P. J., Van Colen, C., Van Hoey, G., and Widdicombe, S.: A bioturbation classification of European marine infaunal invertebrates, Ecol. Evol., 3, 3958–3985, https://doi.org/10.1002/ece3.769, 2013.
Renaud, P., Tessmann, M., Evenset, A., and Christensen, G.: Benthic food-web structure of an Arctic fjord (Kongsfjorden, Svalbard), Mar. Biol. Res., 7, 13–26, https://doi.org/10.1080/17451001003671597, 2011.
Rosenberg, R.: Benthic marine fauna structured by hydrodynamic procesess and food availability, Neth. J. Sea. Res., 34, 303–317, 1995.
Schleuter, D., Daufresne, M., Massol, F., and Argillier, C.: User's guide to functional diversity indices, Ecol. Monogr., 80, 448–469, https://doi.org/10.1890/08-2225.1, 2010.
Solan, M., Aspden, R. J., and Paterson, D. M. (Eds.): Marine biodiversity & ecosystem functioning, 1st Edn., Oxford University Press, Oxford, 2012.
Spitz, J., Ridoux, V., and Brind'Amour, A.: Let's go beyond taxonomy in diet description: Testing a trait-based approach to prey-predator relationships, J. Anim. Ecol., 83, 1137–1148, https://doi.org/10.1111/1365-2656.12218, 2014.
Törnroos, A. and Bonsdorff, E.: Developing the multitrait concept for functional diversity: Lessons from a system rich in functions but poor in species, Ecol. Appl., 22, 2221–2236, https://doi.org/10.1890/11-2042.1, 2012.
Tyler, E. H. M., Somerfield, P. J., Berghe, E. Vanden, Bremner, J., Jackson, E., Langmead, O., Palomares, M. L. D., and Webb, T. J.: Extensive gaps and biases in our knowledge of a well-known fauna: Implications for integrating biological traits into macroecology, Global Ecol. Biogeogr., 21, 922–934, https://doi.org/10.1111/j.1466-8238.2011.00726.x, 2012.
van der Linden, P., Marchini, A., Dolbeth, M., Patrício, J., Veríssimo, H., and Marques, J. C.: The performance of trait-based indices in an estuarine environment, Ecol. Indic., 61, 378–389, https://doi.org/10.1016/j.ecolind.2015.09.039, 2016.
Wassmann, P., Duarte, C. M., Agustí, S., and Sejr, M. K.: Footprints of climate change in the Arctic marine ecosystem, Glob. Change Biol., 17, 1235–1249, https://doi.org/10.1111/j.1365-2486.2010.02311.x, 2011.
Weigel, B., Blenckner, T., and Bonsdorff, E.: Maintained functional diversity in benthic communities in spite of diverging functional identities, Oikos, 125, 1421–1433, https://doi.org/10.1111/oik.02894, 2016.
Weslawski, J., Wlodarska-Kowalczuk, M., and Legezynska, J.: Occurrence of soft bottom macrofauna along the depth gradient in High Arctic, 79 N, Polar Res., 24, 73–88, 2003.
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Döring, M., Giovanni, R., Robertson, T., and Vieglais, D.: Darwin core: An evolving community-developed biodiversity data standard, LoS ONE, 7, e29715, https://doi.org/10.1371/journal.pone.0029715, 2012.
Wiedmann, M., Aschan, M., Certain, G., Dolgov, A., Greenacre, M., Johannesen, E., Planque, B., and Primicerio, R.: Functional diversity of the Barents Sea fish community, Mar. Ecol. Prog. Ser., 495, 205–218, https://doi.org/10.3354/meps10558, 2014.
WoRMS Editorial Board: World Register of Marine Species: available at: http://www.marinespecies.org/ (last access: 20 February 2019), https://doi.org/10.14284/170, 2017.